Chapter 5 Flux Measurements & Inter-Operability
Estimated Time: 3 hours
Course participants: As you review this information, please consider the final course project that you will work on at the over this semester. At the end of this section, you will document an initial research question or idea and associated data needed to address that question, that you may want to explore while pursuing this course.
5.1 Learning Objectives
At the end of this activity, you will be able to:
Define the eddy covariance method
Articulate various carbon storage and flux terms
Understand the structure of bundled eddy covariance data
Be able to process NEON flux data such that it is comparable with AmeriFlux
5.1.1 Guest Lectures
5.1.2 Assignments in this chapter
- NEON Flux Coding Lab Use the NEON flux workflow described in this chapter to merge data from NEON and AmeriFlux (5.7).
5.4 Examples of Other Flux Networks: AMERIFLUX & FLUXNET
AmeriFlux is a network of PI-managed sites measuring ecosystem CO2, water, and energy fluxes in North, Central and South America. It was established to connect research on field sites representing major climate and ecological biomes, including tundra, grasslands, savanna, crops, and conifer, deciduous, and tropical forests. As a grassroots, investigator-driven network, the AmeriFlux community has tailored instrumentation to suit each unique ecosystem. This “coalition of the willing” is diverse in its interests, use of technologies and collaborative approaches. As a result, the AmeriFlux Network continually pioneers new ground.
The network was launched in 1996, after an international workshop on flux measurements in La Thuile, Italy, in 1995, where some of the first year-long flux measurements were presented. Early support for the network came from many sources, including the U.S. Department of Energy’s Terrestrial Carbon Program, the DOE’s National Institute of Global Environmental Change (NIGEC), NASA, NOAA and the US Forest Service. The network grew from about 15 sites in 1997 to more than 110 active sites registered today. Sixty-one other sites, now inactive, have flux data stored in the network’s database. In 2012, the U.S. DOE established the AmeriFlux Management Project (AMP) at Lawrence Berkeley National Laboratory (LBNL) to support the broad AmeriFlux community and the AmeriFlux sites.
View the AMERIFLUX Network-at-a-Glance
AmeriFlux is now one of the DOE Office of Biological and Environmental Research’s (BER) best-known and most highly regarded brands in climate and ecological research. AmeriFlux datasets, and the understanding derived from them, provide crucial linkages between terrestrial ecosystem processes and climate-relevant responses at landscape, regional, and continental scales.
5.5 The Power of Networked Ecology: Bridging to AMERIFLUX and Beyond
Given that AmeriFlux has been collecting and coordinating eddy covariance data across the Americas since 1996. The network provides a common platform for data sharing and collaboration for organizations and individual private investigators collecting flux tower data. There are now >470 registered flux tower sites in North, Central, and South America in the AmeriFlux network, many operated by individual researchers or universities. The towers collect eddy covariance data across a broad range of climate zones and ecosystem types, from Chile to Alaska and everywhere in between.
Now, data from the NEON project is available through the AmeriFlux data portal. The NEON team has formatted data from the NEON flux towers to make it fully compatible with AmeriFlux data. This allows researchers to view, download and analyze data from the NEON flux towers alongside data from all of the other flux towers in the AmeriFlux network.
With 47 flux towers at terrestrial field sites across the U.S., the NEON program is now the largest single contributor of flux tower data to the AmeriFlux network. NEON field sites are located in 20 ecoclimatic zones across the U.S., representing many distinct ecosystems. Eddy covariance data will be served using the same methods at each site for the entire 30-year life of the Observatory, allowing for unprecedented comparability across both time and space.
5.6 Hands On: Introduction to working with NEON eddy flux data
5.6.1 Setup
Start by installing and loading packages and setting options. To work
with the NEON flux data, we need the rhdf5 package, which is hosted on
Bioconductor, and requires a different installation process than CRAN
packages:
install.packages('BiocManager')
BiocManager::install('rhdf5')options(stringsAsFactors=F)
library(neonUtilities)Use the zipsByProduct() function from the neonUtilities package to
download flux data from two sites and two months. The transformations
and functions below will work on any time range and site(s), but two
sites and two months allows us to see all the available functionality
while minimizing download size.
Inputs to the zipsByProduct() function:
dpID: DP4.00200.001, the bundled eddy covariance productpackage: basic (the expanded package is not covered in this tutorial)site: NIWO = Niwot Ridge and HARV = Harvard Foreststartdate: 2018-06 (both dates are inclusive)enddate: 2018-07 (both dates are inclusive)savepath: modify this to something logical on your machinecheck.size: T if you want to see file size before downloading, otherwise F
The download may take a while, especially if you’re on a slow network.
zipsByProduct(dpID="DP4.00200.001", package="basic",
site=c("NIWO", "HARV"),
startdate="2018-06", enddate="2018-07",
savepath="./data",
check.size=F)## Finding available files
##
|
| | 0%
|
|================== | 25%
|
|=================================== | 50%
|
|==================================================== | 75%
|
|======================================================================| 100%
##
## Downloading files totaling approximately 345.260177 MB## ./data/filesToStack00200 already exists. Download will proceed, but check for duplicate files.## Downloading 4 files
##
|
| | 0%
|
|======================= | 33%
|
|=============================================== | 67%
|
|======================================================================| 100%
## 4 files successfully downloaded to ./data/filesToStack002005.6.2 Data Levels
There are five levels of data contained in the eddy flux bundle. For full details, refer to the NEON algorithm document.
Briefly, the data levels are:
- Level 0’ (dp0p): Calibrated raw observations
- Level 1 (dp01): Time-aggregated observations, e.g. 30-minute mean gas concentrations
- Level 2 (dp02): Time-interpolated data, e.g. rate of change of a gas concentration
- Level 3 (dp03): Spatially interpolated data, i.e. vertical profiles
- Level 4 (dp04): Fluxes
The dp0p data are available in the expanded data package and are beyond the scope of this tutorial.
The dp02 and dp03 data are used in storage calculations, and the dp04 data include both the storage and turbulent components. Since many users will want to focus on the net flux data, we’ll start there.
5.6.3 Extract Level 4 data (Fluxes!)
To extract the Level 4 data from the HDF5 files and merge them into a
single table, we’ll use the stackEddy() function from the
neonUtilities package.
stackEddy() requires two inputs:
filepath: Path to a file or folder, which can be any one of:- A zip file of eddy flux data downloaded from the NEON data portal
- A folder of eddy flux data downloaded by the
zipsByProduct()function - The folder of files resulting from unzipping either of 1 or 2
- A single HDF5 file of NEON eddy flux data
level: dp01-4
Input the filepath you downloaded to using zipsByProduct() earlier,
including the filestoStack00200 folder created by the function, and
dp04:
flux <- stackEddy(filepath="./data/filesToStack00200",
level="dp04")## Extracting data
##
|
| | 0%
|
|================== | 25%
|
|=================================== | 50%
|
|==================================================== | 75%
|
|======================================================================| 100%
## Stacking data tables by month
##
|
| | 0%
|
|== | 2%
|
|=== | 5%
|
|===== | 7%
|
|====== | 9%
|
|======== | 11%
|
|========== | 14%
|
|=========== | 16%
|
|============= | 18%
|
|============== | 20%
|
|================ | 23%
|
|================== | 25%
|
|=================== | 27%
|
|===================== | 30%
|
|====================== | 32%
|
|======================== | 34%
|
|========================= | 36%
|
|=========================== | 39%
|
|============================= | 41%
|
|============================== | 43%
|
|================================ | 45%
|
|================================= | 48%
|
|=================================== | 50%
|
|===================================== | 52%
|
|====================================== | 55%
|
|======================================== | 57%
|
|========================================= | 59%
|
|=========================================== | 61%
|
|============================================= | 64%
|
|============================================== | 66%
|
|================================================ | 68%
|
|================================================= | 70%
|
|=================================================== | 73%
|
|==================================================== | 75%
|
|====================================================== | 77%
|
|======================================================== | 80%
|
|========================================================= | 82%
|
|=========================================================== | 84%
|
|============================================================ | 86%
|
|============================================================== | 89%
|
|================================================================ | 91%
|
|================================================================= | 93%
|
|=================================================================== | 95%
|
|==================================================================== | 98%
|
|======================================================================| 100%
## Joining data variables
##
|
| | 0%
|
|== | 2%
|
|=== | 5%
|
|===== | 7%
|
|====== | 9%
|
|======== | 11%
|
|========== | 14%
|
|=========== | 16%
|
|============= | 18%
|
|============== | 20%
|
|================ | 23%
|
|================== | 25%
|
|=================== | 27%
|
|===================== | 30%
|
|====================== | 32%
|
|======================== | 34%
|
|========================= | 36%
|
|=========================== | 39%
|
|============================= | 41%
|
|============================== | 43%
|
|================================ | 45%
|
|================================= | 48%
|
|=================================== | 50%
|
|===================================== | 52%
|
|====================================== | 55%
|
|======================================== | 57%
|
|========================================= | 59%
|
|=========================================== | 61%
|
|============================================= | 64%
|
|============================================== | 66%
|
|================================================ | 68%
|
|================================================= | 70%
|
|=================================================== | 73%
|
|==================================================== | 75%
|
|====================================================== | 77%
|
|======================================================== | 80%
|
|========================================================= | 82%
|
|=========================================================== | 84%
|
|============================================================ | 86%
|
|============================================================== | 89%
|
|================================================================ | 91%
|
|================================================================= | 93%
|
|=================================================================== | 95%
|
|==================================================================== | 98%
|
|======================================================================| 100%We now have an object called flux. It’s a named list containing four
tables: one table for each site’s data, and variables and objDesc
tables.
names(flux)## [1] "HARV" "NIWO" "variables" "objDesc" "issueLog"Let’s look at the contents of one of the site data files:
knitr::kable(head(flux$NIWO))| timeBgn | timeEnd | data.fluxCo2.nsae.flux | data.fluxCo2.stor.flux | data.fluxCo2.turb.flux | data.fluxH2o.nsae.flux | data.fluxH2o.stor.flux | data.fluxH2o.turb.flux | data.fluxMome.turb.veloFric | data.fluxTemp.nsae.flux | data.fluxTemp.stor.flux | data.fluxTemp.turb.flux | data.foot.stat.angZaxsErth | data.foot.stat.distReso | data.foot.stat.veloYaxsHorSd | data.foot.stat.veloZaxsHorSd | data.foot.stat.veloFric | data.foot.stat.distZaxsMeasDisp | data.foot.stat.distZaxsRgh | data.foot.stat.distZaxsAbl | data.foot.stat.distXaxs90 | data.foot.stat.distXaxsMax | data.foot.stat.distYaxs90 | qfqm.fluxCo2.nsae.qfFinl | qfqm.fluxCo2.stor.qfFinl | qfqm.fluxCo2.turb.qfFinl | qfqm.fluxH2o.nsae.qfFinl | qfqm.fluxH2o.stor.qfFinl | qfqm.fluxH2o.turb.qfFinl | qfqm.fluxMome.turb.qfFinl | qfqm.fluxTemp.nsae.qfFinl | qfqm.fluxTemp.stor.qfFinl | qfqm.fluxTemp.turb.qfFinl | qfqm.foot.turb.qfFinl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2018-06-01 00:00:00 | 2018-06-01 00:29:59 | 0.1713858 | -0.0634816 | 0.2348674 | 15.876622 | 3.3334970 | 12.543125 | 0.2047081 | 4.7565505 | -1.4575094 | 6.2140599 | 94.2262 | 8.34 | 0.7955893 | 0.2713232 | 0.2025427 | 8.34 | 0.0410571 | 1000 | 325.26 | 133.44 | 25.02 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| 2018-06-01 00:30:00 | 2018-06-01 00:59:59 | 0.9251711 | 0.0874815 | 0.8376896 | 8.089274 | -1.2063258 | 9.295600 | 0.1923735 | -0.2717454 | 0.3403877 | -0.6121331 | 355.4252 | 8.34 | 0.8590177 | 0.2300000 | 0.2000000 | 8.34 | 0.2799194 | 1000 | 266.88 | 108.42 | 50.04 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| 2018-06-01 01:00:00 | 2018-06-01 01:29:59 | 0.5005812 | 0.0223170 | 0.4782642 | 5.290594 | -4.4190781 | 9.709672 | 0.1200918 | -4.2055147 | 0.1870677 | -4.3925824 | 359.8013 | 8.34 | 1.2601763 | 0.2300000 | 0.2000000 | 8.34 | 0.2129323 | 1000 | 275.22 | 116.76 | 66.72 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 2018-06-01 01:30:00 | 2018-06-01 01:59:59 | 0.8032820 | 0.2556931 | 0.5475889 | 9.190214 | 0.2030371 | 8.987177 | 0.1177545 | -13.3834484 | -2.4904300 | -10.8930185 | 137.7743 | 8.34 | 0.7332641 | 0.2300000 | 0.2000000 | 8.34 | 0.8340000 | 1000 | 208.50 | 83.40 | 75.06 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 2018-06-01 02:00:00 | 2018-06-01 02:29:59 | 0.4897685 | 0.2309047 | 0.2588638 | 3.111909 | 0.1349363 | 2.976973 | 0.1589189 | -5.1854815 | -0.7514531 | -4.4340284 | 188.4799 | 8.34 | 0.7096286 | 0.2300000 | 0.2000000 | 8.34 | 0.8340000 | 1000 | 208.50 | 83.40 | 66.72 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2018-06-01 02:30:00 | 2018-06-01 02:59:59 | 0.9223979 | 0.0622858 | 0.8601121 | 4.613676 | -0.3929445 | 5.006621 | 0.1114406 | -7.7365481 | -1.9046775 | -5.8318707 | 183.1920 | 8.34 | 0.3789859 | 0.2300000 | 0.2000000 | 8.34 | 0.8340000 | 1000 | 208.50 | 83.40 | 41.70 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
The variables and objDesc tables can help you interpret the column
headers in the data table. The objDesc table contains definitions for
many of the terms used in the eddy flux data product, but it isn’t
complete. To get the terms of interest, we’ll break up the column
headers into individual terms and look for them in the objDesc table:
term <- unlist(strsplit(names(flux$NIWO), split=".", fixed=T))
flux$objDesc[which(flux$objDesc$Object %in% term),]## Object
## 138 angZaxsErth
## 171 data
## 343 qfFinl
## 420 qfqm
## 604 timeBgn
## 605 timeEnd
## Description
## 138 Wind direction
## 171 Represents data fields
## 343 The final quality flag indicating if the data are valid for the given aggregation period (1=fail, 0=pass)
## 420 Quality flag and quality metrics, represents quality flags and quality metrics that accompany the provided data
## 604 The beginning time of the aggregation period
## 605 The end time of the aggregation periodknitr::kable(term)| x |
|---|
| timeBgn |
| timeEnd |
| data |
| fluxCo2 |
| nsae |
| flux |
| data |
| fluxCo2 |
| stor |
| flux |
| data |
| fluxCo2 |
| turb |
| flux |
| data |
| fluxH2o |
| nsae |
| flux |
| data |
| fluxH2o |
| stor |
| flux |
| data |
| fluxH2o |
| turb |
| flux |
| data |
| fluxMome |
| turb |
| veloFric |
| data |
| fluxTemp |
| nsae |
| flux |
| data |
| fluxTemp |
| stor |
| flux |
| data |
| fluxTemp |
| turb |
| flux |
| data |
| foot |
| stat |
| angZaxsErth |
| data |
| foot |
| stat |
| distReso |
| data |
| foot |
| stat |
| veloYaxsHorSd |
| data |
| foot |
| stat |
| veloZaxsHorSd |
| data |
| foot |
| stat |
| veloFric |
| data |
| foot |
| stat |
| distZaxsMeasDisp |
| data |
| foot |
| stat |
| distZaxsRgh |
| data |
| foot |
| stat |
| distZaxsAbl |
| data |
| foot |
| stat |
| distXaxs90 |
| data |
| foot |
| stat |
| distXaxsMax |
| data |
| foot |
| stat |
| distYaxs90 |
| qfqm |
| fluxCo2 |
| nsae |
| qfFinl |
| qfqm |
| fluxCo2 |
| stor |
| qfFinl |
| qfqm |
| fluxCo2 |
| turb |
| qfFinl |
| qfqm |
| fluxH2o |
| nsae |
| qfFinl |
| qfqm |
| fluxH2o |
| stor |
| qfFinl |
| qfqm |
| fluxH2o |
| turb |
| qfFinl |
| qfqm |
| fluxMome |
| turb |
| qfFinl |
| qfqm |
| fluxTemp |
| nsae |
| qfFinl |
| qfqm |
| fluxTemp |
| stor |
| qfFinl |
| qfqm |
| fluxTemp |
| turb |
| qfFinl |
| qfqm |
| foot |
| turb |
| qfFinl |
For the terms that aren’t captured here, fluxCo2, fluxH2o, and
fluxTemp are self-explanatory. The flux components are
turb: Turbulent fluxstor: Storagensae: Net surface-atmosphere exchange
The variables table contains the units for each field:
knitr::kable(flux$variables)| category | system | variable | stat | units |
|---|---|---|---|---|
| data | fluxCo2 | nsae | timeBgn | NA |
| data | fluxCo2 | nsae | timeEnd | NA |
| data | fluxCo2 | nsae | flux | umolCo2 m-2 s-1 |
| data | fluxCo2 | stor | timeBgn | NA |
| data | fluxCo2 | stor | timeEnd | NA |
| data | fluxCo2 | stor | flux | umolCo2 m-2 s-1 |
| data | fluxCo2 | turb | timeBgn | NA |
| data | fluxCo2 | turb | timeEnd | NA |
| data | fluxCo2 | turb | flux | umolCo2 m-2 s-1 |
| data | fluxH2o | nsae | timeBgn | NA |
| data | fluxH2o | nsae | timeEnd | NA |
| data | fluxH2o | nsae | flux | W m-2 |
| data | fluxH2o | stor | timeBgn | NA |
| data | fluxH2o | stor | timeEnd | NA |
| data | fluxH2o | stor | flux | W m-2 |
| data | fluxH2o | turb | timeBgn | NA |
| data | fluxH2o | turb | timeEnd | NA |
| data | fluxH2o | turb | flux | W m-2 |
| data | fluxMome | turb | timeBgn | NA |
| data | fluxMome | turb | timeEnd | NA |
| data | fluxMome | turb | veloFric | m s-1 |
| data | fluxTemp | nsae | timeBgn | NA |
| data | fluxTemp | nsae | timeEnd | NA |
| data | fluxTemp | nsae | flux | W m-2 |
| data | fluxTemp | stor | timeBgn | NA |
| data | fluxTemp | stor | timeEnd | NA |
| data | fluxTemp | stor | flux | W m-2 |
| data | fluxTemp | turb | timeBgn | NA |
| data | fluxTemp | turb | timeEnd | NA |
| data | fluxTemp | turb | flux | W m-2 |
| data | foot | stat | timeBgn | NA |
| data | foot | stat | timeEnd | NA |
| data | foot | stat | angZaxsErth | deg |
| data | foot | stat | distReso | m |
| data | foot | stat | veloYaxsHorSd | m s-1 |
| data | foot | stat | veloZaxsHorSd | m s-1 |
| data | foot | stat | veloFric | m s-1 |
| data | foot | stat | distZaxsMeasDisp | m |
| data | foot | stat | distZaxsRgh | m |
| data | foot | stat | distZaxsAbl | m |
| data | foot | stat | distXaxs90 | m |
| data | foot | stat | distXaxsMax | m |
| data | foot | stat | distYaxs90 | m |
| qfqm | fluxCo2 | nsae | timeBgn | NA |
| qfqm | fluxCo2 | nsae | timeEnd | NA |
| qfqm | fluxCo2 | nsae | qfFinl | NA |
| qfqm | fluxCo2 | stor | qfFinl | NA |
| qfqm | fluxCo2 | stor | timeBgn | NA |
| qfqm | fluxCo2 | stor | timeEnd | NA |
| qfqm | fluxCo2 | turb | timeBgn | NA |
| qfqm | fluxCo2 | turb | timeEnd | NA |
| qfqm | fluxCo2 | turb | qfFinl | NA |
| qfqm | fluxH2o | nsae | timeBgn | NA |
| qfqm | fluxH2o | nsae | timeEnd | NA |
| qfqm | fluxH2o | nsae | qfFinl | NA |
| qfqm | fluxH2o | stor | qfFinl | NA |
| qfqm | fluxH2o | stor | timeBgn | NA |
| qfqm | fluxH2o | stor | timeEnd | NA |
| qfqm | fluxH2o | turb | timeBgn | NA |
| qfqm | fluxH2o | turb | timeEnd | NA |
| qfqm | fluxH2o | turb | qfFinl | NA |
| qfqm | fluxMome | turb | timeBgn | NA |
| qfqm | fluxMome | turb | timeEnd | NA |
| qfqm | fluxMome | turb | qfFinl | NA |
| qfqm | fluxTemp | nsae | timeBgn | NA |
| qfqm | fluxTemp | nsae | timeEnd | NA |
| qfqm | fluxTemp | nsae | qfFinl | NA |
| qfqm | fluxTemp | stor | qfFinl | NA |
| qfqm | fluxTemp | stor | timeBgn | NA |
| qfqm | fluxTemp | stor | timeEnd | NA |
| qfqm | fluxTemp | turb | timeBgn | NA |
| qfqm | fluxTemp | turb | timeEnd | NA |
| qfqm | fluxTemp | turb | qfFinl | NA |
| qfqm | foot | turb | timeBgn | NA |
| qfqm | foot | turb | timeEnd | NA |
| qfqm | foot | turb | qfFinl | NA |
Let’s plot some data! First, we’ll need to convert the time stamps to an R date-time format (right now they’re just character fields).
5.6.4 Time stamps
NEON sensor data come with time stamps for both the start and end of the averaging period. Depending on the analysis you’re doing, you may want to use one or the other; for general plotting, re-formatting, and transformations, I prefer to use the start time, because there are some small inconsistencies between data products in a few of the end time stamps.
Note that all NEON data use UTC time, noted as tz="GMT" in the
code below. This is true across NEON’s instrumented, observational, and
airborne measurements. When working with NEON data, it’s best to keep
everything in UTC as much as possible, otherwise it’s very easy to end
up with data in mismatched times, which can cause insidious and
hard-to-detect problems. Be sure to include the tz argument in all the
lines of code below - if there is no time zone specified, R will default
to the local time zone it detects on your operating system.
timeB <- as.POSIXct(flux$NIWO$timeBgn,
format="%Y-%m-%dT%H:%M:%S",
tz="GMT")
flux$NIWO <- cbind(timeB, flux$NIWO)plot(flux$NIWO$data.fluxCo2.nsae.flux~timeB,
pch=".", xlab="Date", ylab="CO2 flux",
xaxt="n")
axis.POSIXct(1, x=timeB, format="%Y-%m-%d")Like a lot of flux data, these data have some stray spikes, but there is a clear diurnal pattern going into the growing season.
Let’s trim down to just two days of data to see a few other details.
plot(flux$NIWO$data.fluxCo2.nsae.flux~timeB,
pch=20, xlab="Date", ylab="CO2 flux",
xlim=c(as.POSIXct("2018-07-07", tz="GMT"),
as.POSIXct("2018-07-09", tz="GMT")),
ylim=c(-20,20), xaxt="n")
axis.POSIXct(1, x=timeB, format="%Y-%m-%d %H:%M:%S")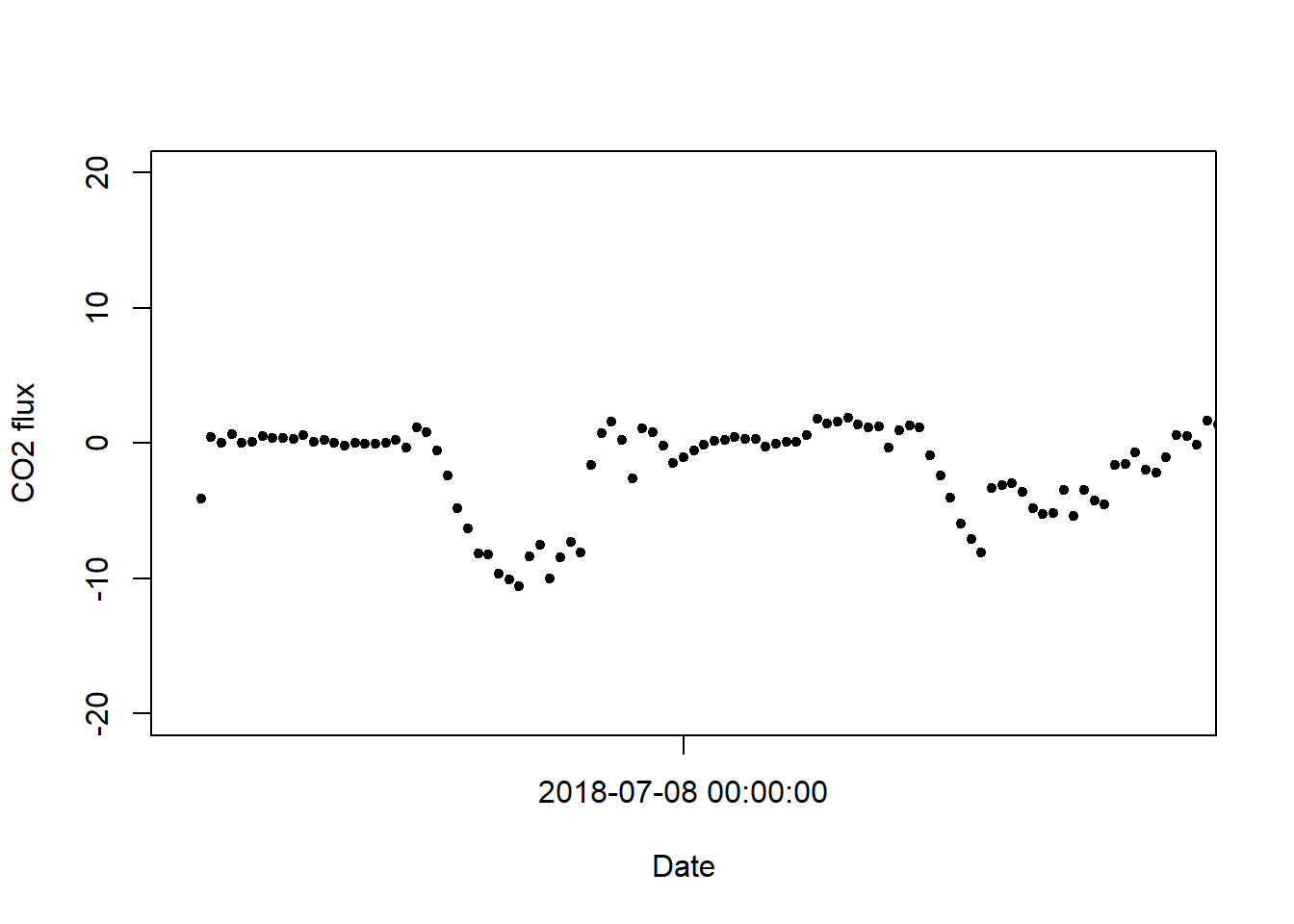
Note the timing of C uptake; the UTC time zone is clear here, where uptake occurs at times that appear to be during the night.
5.6.5 Merge flux data with other sensor data
Many of the data sets we would use to interpret and model flux data are measured as part of the NEON project, but are not present in the eddy flux data product bundle. In this section, we’ll download PAR data and merge them with the flux data; the steps taken here can be applied to any of the NEON instrumented (IS) data products.
5.6.5.1 Download PAR data
To get NEON PAR data, use the loadByProduct() function from the
neonUtilities package. loadByProduct() takes the same inputs as
zipsByProduct(), but it loads the downloaded data directly into the
current R environment.
Let’s download PAR data matching the Niwot Ridge flux data. The inputs needed are:
dpID: DP1.00024.001site: NIWOstartdate: 2018-06enddate: 2018-07package: basicavg: 30
The new input here is avg=30, which downloads only the 30-minute data.
Since the flux data are at a 30-minute resolution, we can save on
download time by disregarding the 1-minute data files (which are of
course 30 times larger). The avg input can be left off if you want to
download all available averaging intervals.
pr <- loadByProduct("DP1.00024.001", site="NIWO", avg=30,
startdate="2018-06", enddate="2018-07",
package="basic", check.size=F)## Input parameter avg is deprecated; use timeIndex to download by time interval.
## Finding available files
##
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
##
## Downloading files totaling approximately 1.304794 MB
## Downloading 11 files
##
|
| | 0%
|
|======= | 10%
|
|============== | 20%
|
|===================== | 30%
|
|============================ | 40%
|
|=================================== | 50%
|
|========================================== | 60%
|
|================================================= | 70%
|
|======================================================== | 80%
|
|=============================================================== | 90%
|
|======================================================================| 100%
##
## Stacking operation across a single core.
## Stacking table PARPAR_30min
## Merged the most recent publication of sensor position files for each site and saved to /stackedFiles
## Copied the most recent publication of variable definition file to /stackedFiles
## Finished: Stacked 1 data tables and 3 metadata tables!
## Stacking took 0.3194668 secspr is another named list, and again, metadata and units can be found
in the variables table. The PARPAR_30min table contains a
verticalPosition field. This field indicates the position on the
tower, with 10 being the first tower level, and 20, 30, etc going up the
tower.
5.6.5.2 Join PAR to flux data
We’ll connect PAR data from the tower top to the flux data.
pr.top <- pr$PARPAR_30min[which(pr$PARPAR_30min$verticalPosition==
max(pr$PARPAR_30min$verticalPosition)),]loadByProduct() automatically converts time stamps when it reads the
data, so here we just need to indicate which time field to use to merge
the flux and PAR data.
timeB <- pr.top$startDateTime
pr.top <- cbind(timeB, pr.top)And merge the two datasets:
fx.pr <- merge(pr.top, flux$NIWO, by="timeB")plot(fx.pr$data.fluxCo2.nsae.flux~fx.pr$PARMean,
pch=".", ylim=c(-20,20),
xlab="PAR", ylab="CO2 flux")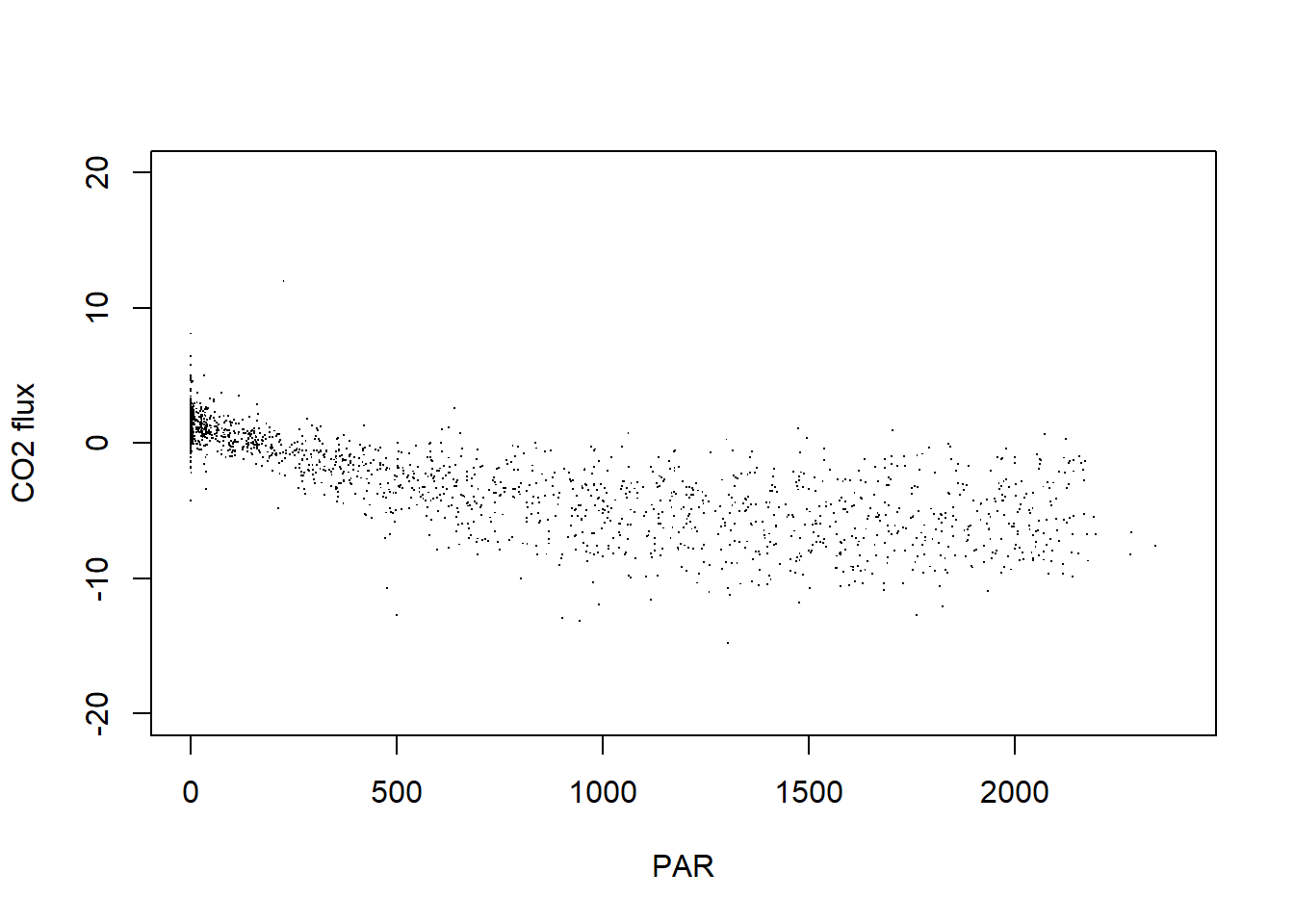
If you’re interested in data in the eddy covariance bundle besides the net flux data, the rest of this tutorial will guide you through how to get those data out of the bundle.
5.6.6 Vertical profile data (Level 3)
The Level 3 (dp03) data are the spatially interpolated profiles of the
rates of change of CO2, H2O, and temperature.
Extract the Level 3 data from the HDF5 file using stackEddy() with the
same syntax as for the Level 4 data.
prof <- stackEddy(filepath="./data/filesToStack00200/",
level="dp03")## Extracting data
##
|
| | 0%
|
|================== | 25%
|
|=================================== | 50%
|
|==================================================== | 75%
|
|======================================================================| 100%
## Stacking data tables by month
##
|
| | 0%
|
|====== | 8%
|
|============ | 17%
|
|================== | 25%
|
|======================= | 33%
|
|============================= | 42%
|
|=================================== | 50%
|
|========================================= | 58%
|
|=============================================== | 67%
|
|==================================================== | 75%
|
|========================================================== | 83%
|
|================================================================ | 92%
|
|======================================================================| 100%
## Joining data variables
##
|
| | 0%
|
|====== | 8%
|
|============ | 17%
|
|================== | 25%
|
|======================= | 33%
|
|============================= | 42%
|
|=================================== | 50%
|
|========================================= | 58%
|
|=============================================== | 67%
|
|==================================================== | 75%
|
|========================================================== | 83%
|
|================================================================ | 92%
|
|======================================================================| 100%knitr::kable(head(prof$NIWO))| timeBgn | timeEnd | data.co2Stor.rateRtioMoleDryCo2.X0.1.m | data.co2Stor.rateRtioMoleDryCo2.X0.2.m | data.co2Stor.rateRtioMoleDryCo2.X0.3.m | data.co2Stor.rateRtioMoleDryCo2.X0.4.m | data.co2Stor.rateRtioMoleDryCo2.X0.5.m | data.co2Stor.rateRtioMoleDryCo2.X0.6.m | data.co2Stor.rateRtioMoleDryCo2.X0.7.m | data.co2Stor.rateRtioMoleDryCo2.X0.8.m | data.co2Stor.rateRtioMoleDryCo2.X0.9.m | data.co2Stor.rateRtioMoleDryCo2.X1.m | data.co2Stor.rateRtioMoleDryCo2.X1.1.m | data.co2Stor.rateRtioMoleDryCo2.X1.2.m | data.co2Stor.rateRtioMoleDryCo2.X1.3.m | data.co2Stor.rateRtioMoleDryCo2.X1.4.m | data.co2Stor.rateRtioMoleDryCo2.X1.5.m | data.co2Stor.rateRtioMoleDryCo2.X1.6.m | data.co2Stor.rateRtioMoleDryCo2.X1.7.m | data.co2Stor.rateRtioMoleDryCo2.X1.8.m | data.co2Stor.rateRtioMoleDryCo2.X1.9.m | data.co2Stor.rateRtioMoleDryCo2.X2.m | data.co2Stor.rateRtioMoleDryCo2.X2.1.m | data.co2Stor.rateRtioMoleDryCo2.X2.2.m | data.co2Stor.rateRtioMoleDryCo2.X2.3.m | data.co2Stor.rateRtioMoleDryCo2.X2.4.m | data.co2Stor.rateRtioMoleDryCo2.X2.5.m | data.co2Stor.rateRtioMoleDryCo2.X2.6.m | data.co2Stor.rateRtioMoleDryCo2.X2.7.m | data.co2Stor.rateRtioMoleDryCo2.X2.8.m | data.co2Stor.rateRtioMoleDryCo2.X2.9.m | data.co2Stor.rateRtioMoleDryCo2.X3.m | data.co2Stor.rateRtioMoleDryCo2.X3.1.m | data.co2Stor.rateRtioMoleDryCo2.X3.2.m | data.co2Stor.rateRtioMoleDryCo2.X3.3.m | data.co2Stor.rateRtioMoleDryCo2.X3.4.m | data.co2Stor.rateRtioMoleDryCo2.X3.5.m | data.co2Stor.rateRtioMoleDryCo2.X3.6.m | data.co2Stor.rateRtioMoleDryCo2.X3.7.m | data.co2Stor.rateRtioMoleDryCo2.X3.8.m | data.co2Stor.rateRtioMoleDryCo2.X3.9.m | data.co2Stor.rateRtioMoleDryCo2.X4.m | data.co2Stor.rateRtioMoleDryCo2.X4.1.m | data.co2Stor.rateRtioMoleDryCo2.X4.2.m | data.co2Stor.rateRtioMoleDryCo2.X4.3.m | data.co2Stor.rateRtioMoleDryCo2.X4.4.m | data.co2Stor.rateRtioMoleDryCo2.X4.5.m | data.co2Stor.rateRtioMoleDryCo2.X4.6.m | data.co2Stor.rateRtioMoleDryCo2.X4.7.m | data.co2Stor.rateRtioMoleDryCo2.X4.8.m | data.co2Stor.rateRtioMoleDryCo2.X4.9.m | data.co2Stor.rateRtioMoleDryCo2.X5.m | data.co2Stor.rateRtioMoleDryCo2.X5.1.m | data.co2Stor.rateRtioMoleDryCo2.X5.2.m | data.co2Stor.rateRtioMoleDryCo2.X5.3.m | data.co2Stor.rateRtioMoleDryCo2.X5.4.m | data.co2Stor.rateRtioMoleDryCo2.X5.5.m | data.co2Stor.rateRtioMoleDryCo2.X5.6.m | data.co2Stor.rateRtioMoleDryCo2.X5.7.m | data.co2Stor.rateRtioMoleDryCo2.X5.8.m | data.co2Stor.rateRtioMoleDryCo2.X5.9.m | data.co2Stor.rateRtioMoleDryCo2.X6.m | data.co2Stor.rateRtioMoleDryCo2.X6.1.m | data.co2Stor.rateRtioMoleDryCo2.X6.2.m | data.co2Stor.rateRtioMoleDryCo2.X6.3.m | data.co2Stor.rateRtioMoleDryCo2.X6.4.m | data.co2Stor.rateRtioMoleDryCo2.X6.5.m | data.co2Stor.rateRtioMoleDryCo2.X6.6.m | data.co2Stor.rateRtioMoleDryCo2.X6.7.m | data.co2Stor.rateRtioMoleDryCo2.X6.8.m | data.co2Stor.rateRtioMoleDryCo2.X6.9.m | data.co2Stor.rateRtioMoleDryCo2.X7.m | data.co2Stor.rateRtioMoleDryCo2.X7.1.m | data.co2Stor.rateRtioMoleDryCo2.X7.2.m | data.co2Stor.rateRtioMoleDryCo2.X7.3.m | data.co2Stor.rateRtioMoleDryCo2.X7.4.m | data.co2Stor.rateRtioMoleDryCo2.X7.5.m | data.co2Stor.rateRtioMoleDryCo2.X7.6.m | data.co2Stor.rateRtioMoleDryCo2.X7.7.m | data.co2Stor.rateRtioMoleDryCo2.X7.8.m | data.co2Stor.rateRtioMoleDryCo2.X7.9.m | data.co2Stor.rateRtioMoleDryCo2.X8.m | data.co2Stor.rateRtioMoleDryCo2.X8.1.m | data.co2Stor.rateRtioMoleDryCo2.X8.2.m | data.co2Stor.rateRtioMoleDryCo2.X8.3.m | data.co2Stor.rateRtioMoleDryCo2.X8.4.m | data.h2oStor.rateRtioMoleDryH2o.X0.1.m | data.h2oStor.rateRtioMoleDryH2o.X0.2.m | data.h2oStor.rateRtioMoleDryH2o.X0.3.m | data.h2oStor.rateRtioMoleDryH2o.X0.4.m | data.h2oStor.rateRtioMoleDryH2o.X0.5.m | data.h2oStor.rateRtioMoleDryH2o.X0.6.m | data.h2oStor.rateRtioMoleDryH2o.X0.7.m | data.h2oStor.rateRtioMoleDryH2o.X0.8.m | data.h2oStor.rateRtioMoleDryH2o.X0.9.m | data.h2oStor.rateRtioMoleDryH2o.X1.m | data.h2oStor.rateRtioMoleDryH2o.X1.1.m | data.h2oStor.rateRtioMoleDryH2o.X1.2.m | data.h2oStor.rateRtioMoleDryH2o.X1.3.m | data.h2oStor.rateRtioMoleDryH2o.X1.4.m | data.h2oStor.rateRtioMoleDryH2o.X1.5.m | data.h2oStor.rateRtioMoleDryH2o.X1.6.m | data.h2oStor.rateRtioMoleDryH2o.X1.7.m | data.h2oStor.rateRtioMoleDryH2o.X1.8.m | data.h2oStor.rateRtioMoleDryH2o.X1.9.m | data.h2oStor.rateRtioMoleDryH2o.X2.m | data.h2oStor.rateRtioMoleDryH2o.X2.1.m | data.h2oStor.rateRtioMoleDryH2o.X2.2.m | data.h2oStor.rateRtioMoleDryH2o.X2.3.m | data.h2oStor.rateRtioMoleDryH2o.X2.4.m | data.h2oStor.rateRtioMoleDryH2o.X2.5.m | data.h2oStor.rateRtioMoleDryH2o.X2.6.m | data.h2oStor.rateRtioMoleDryH2o.X2.7.m | data.h2oStor.rateRtioMoleDryH2o.X2.8.m | data.h2oStor.rateRtioMoleDryH2o.X2.9.m | data.h2oStor.rateRtioMoleDryH2o.X3.m | data.h2oStor.rateRtioMoleDryH2o.X3.1.m | data.h2oStor.rateRtioMoleDryH2o.X3.2.m | data.h2oStor.rateRtioMoleDryH2o.X3.3.m | data.h2oStor.rateRtioMoleDryH2o.X3.4.m | data.h2oStor.rateRtioMoleDryH2o.X3.5.m | data.h2oStor.rateRtioMoleDryH2o.X3.6.m | data.h2oStor.rateRtioMoleDryH2o.X3.7.m | data.h2oStor.rateRtioMoleDryH2o.X3.8.m | data.h2oStor.rateRtioMoleDryH2o.X3.9.m | data.h2oStor.rateRtioMoleDryH2o.X4.m | data.h2oStor.rateRtioMoleDryH2o.X4.1.m | data.h2oStor.rateRtioMoleDryH2o.X4.2.m | data.h2oStor.rateRtioMoleDryH2o.X4.3.m | data.h2oStor.rateRtioMoleDryH2o.X4.4.m | data.h2oStor.rateRtioMoleDryH2o.X4.5.m | data.h2oStor.rateRtioMoleDryH2o.X4.6.m | data.h2oStor.rateRtioMoleDryH2o.X4.7.m | data.h2oStor.rateRtioMoleDryH2o.X4.8.m | data.h2oStor.rateRtioMoleDryH2o.X4.9.m | data.h2oStor.rateRtioMoleDryH2o.X5.m | data.h2oStor.rateRtioMoleDryH2o.X5.1.m | data.h2oStor.rateRtioMoleDryH2o.X5.2.m | data.h2oStor.rateRtioMoleDryH2o.X5.3.m | data.h2oStor.rateRtioMoleDryH2o.X5.4.m | data.h2oStor.rateRtioMoleDryH2o.X5.5.m | data.h2oStor.rateRtioMoleDryH2o.X5.6.m | data.h2oStor.rateRtioMoleDryH2o.X5.7.m | data.h2oStor.rateRtioMoleDryH2o.X5.8.m | data.h2oStor.rateRtioMoleDryH2o.X5.9.m | data.h2oStor.rateRtioMoleDryH2o.X6.m | data.h2oStor.rateRtioMoleDryH2o.X6.1.m | data.h2oStor.rateRtioMoleDryH2o.X6.2.m | data.h2oStor.rateRtioMoleDryH2o.X6.3.m | data.h2oStor.rateRtioMoleDryH2o.X6.4.m | data.h2oStor.rateRtioMoleDryH2o.X6.5.m | data.h2oStor.rateRtioMoleDryH2o.X6.6.m | data.h2oStor.rateRtioMoleDryH2o.X6.7.m | data.h2oStor.rateRtioMoleDryH2o.X6.8.m | data.h2oStor.rateRtioMoleDryH2o.X6.9.m | data.h2oStor.rateRtioMoleDryH2o.X7.m | data.h2oStor.rateRtioMoleDryH2o.X7.1.m | data.h2oStor.rateRtioMoleDryH2o.X7.2.m | data.h2oStor.rateRtioMoleDryH2o.X7.3.m | data.h2oStor.rateRtioMoleDryH2o.X7.4.m | data.h2oStor.rateRtioMoleDryH2o.X7.5.m | data.h2oStor.rateRtioMoleDryH2o.X7.6.m | data.h2oStor.rateRtioMoleDryH2o.X7.7.m | data.h2oStor.rateRtioMoleDryH2o.X7.8.m | data.h2oStor.rateRtioMoleDryH2o.X7.9.m | data.h2oStor.rateRtioMoleDryH2o.X8.m | data.h2oStor.rateRtioMoleDryH2o.X8.1.m | data.h2oStor.rateRtioMoleDryH2o.X8.2.m | data.h2oStor.rateRtioMoleDryH2o.X8.3.m | data.h2oStor.rateRtioMoleDryH2o.X8.4.m | data.tempStor.rateTemp.X0.1.m | data.tempStor.rateTemp.X0.2.m | data.tempStor.rateTemp.X0.3.m | data.tempStor.rateTemp.X0.4.m | data.tempStor.rateTemp.X0.5.m | data.tempStor.rateTemp.X0.6.m | data.tempStor.rateTemp.X0.7.m | data.tempStor.rateTemp.X0.8.m | data.tempStor.rateTemp.X0.9.m | data.tempStor.rateTemp.X1.m | data.tempStor.rateTemp.X1.1.m | data.tempStor.rateTemp.X1.2.m | data.tempStor.rateTemp.X1.3.m | data.tempStor.rateTemp.X1.4.m | data.tempStor.rateTemp.X1.5.m | data.tempStor.rateTemp.X1.6.m | data.tempStor.rateTemp.X1.7.m | data.tempStor.rateTemp.X1.8.m | data.tempStor.rateTemp.X1.9.m | data.tempStor.rateTemp.X2.m | data.tempStor.rateTemp.X2.1.m | data.tempStor.rateTemp.X2.2.m | data.tempStor.rateTemp.X2.3.m | data.tempStor.rateTemp.X2.4.m | data.tempStor.rateTemp.X2.5.m | data.tempStor.rateTemp.X2.6.m | data.tempStor.rateTemp.X2.7.m | data.tempStor.rateTemp.X2.8.m | data.tempStor.rateTemp.X2.9.m | data.tempStor.rateTemp.X3.m | data.tempStor.rateTemp.X3.1.m | data.tempStor.rateTemp.X3.2.m | data.tempStor.rateTemp.X3.3.m | data.tempStor.rateTemp.X3.4.m | data.tempStor.rateTemp.X3.5.m | data.tempStor.rateTemp.X3.6.m | data.tempStor.rateTemp.X3.7.m | data.tempStor.rateTemp.X3.8.m | data.tempStor.rateTemp.X3.9.m | data.tempStor.rateTemp.X4.m | data.tempStor.rateTemp.X4.1.m | data.tempStor.rateTemp.X4.2.m | data.tempStor.rateTemp.X4.3.m | data.tempStor.rateTemp.X4.4.m | data.tempStor.rateTemp.X4.5.m | data.tempStor.rateTemp.X4.6.m | data.tempStor.rateTemp.X4.7.m | data.tempStor.rateTemp.X4.8.m | data.tempStor.rateTemp.X4.9.m | data.tempStor.rateTemp.X5.m | data.tempStor.rateTemp.X5.1.m | data.tempStor.rateTemp.X5.2.m | data.tempStor.rateTemp.X5.3.m | data.tempStor.rateTemp.X5.4.m | data.tempStor.rateTemp.X5.5.m | data.tempStor.rateTemp.X5.6.m | data.tempStor.rateTemp.X5.7.m | data.tempStor.rateTemp.X5.8.m | data.tempStor.rateTemp.X5.9.m | data.tempStor.rateTemp.X6.m | data.tempStor.rateTemp.X6.1.m | data.tempStor.rateTemp.X6.2.m | data.tempStor.rateTemp.X6.3.m | data.tempStor.rateTemp.X6.4.m | data.tempStor.rateTemp.X6.5.m | data.tempStor.rateTemp.X6.6.m | data.tempStor.rateTemp.X6.7.m | data.tempStor.rateTemp.X6.8.m | data.tempStor.rateTemp.X6.9.m | data.tempStor.rateTemp.X7.m | data.tempStor.rateTemp.X7.1.m | data.tempStor.rateTemp.X7.2.m | data.tempStor.rateTemp.X7.3.m | data.tempStor.rateTemp.X7.4.m | data.tempStor.rateTemp.X7.5.m | data.tempStor.rateTemp.X7.6.m | data.tempStor.rateTemp.X7.7.m | data.tempStor.rateTemp.X7.8.m | data.tempStor.rateTemp.X7.9.m | data.tempStor.rateTemp.X8.m | data.tempStor.rateTemp.X8.1.m | data.tempStor.rateTemp.X8.2.m | data.tempStor.rateTemp.X8.3.m | data.tempStor.rateTemp.X8.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X0.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X1.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X2.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X3.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X4.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X5.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X6.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.4.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.5.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.6.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.7.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X7.9.m | qfqm.co2Stor.rateRtioMoleDryCo2.X8.m | qfqm.co2Stor.rateRtioMoleDryCo2.X8.1.m | qfqm.co2Stor.rateRtioMoleDryCo2.X8.2.m | qfqm.co2Stor.rateRtioMoleDryCo2.X8.3.m | qfqm.co2Stor.rateRtioMoleDryCo2.X8.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X0.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X1.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X2.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X3.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X4.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X5.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X6.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.4.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.5.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.6.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.7.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X7.9.m | qfqm.h2oStor.rateRtioMoleDryH2o.X8.m | qfqm.h2oStor.rateRtioMoleDryH2o.X8.1.m | qfqm.h2oStor.rateRtioMoleDryH2o.X8.2.m | qfqm.h2oStor.rateRtioMoleDryH2o.X8.3.m | qfqm.h2oStor.rateRtioMoleDryH2o.X8.4.m | qfqm.tempStor.rateTemp.X0.1.m | qfqm.tempStor.rateTemp.X0.2.m | qfqm.tempStor.rateTemp.X0.3.m | qfqm.tempStor.rateTemp.X0.4.m | qfqm.tempStor.rateTemp.X0.5.m | qfqm.tempStor.rateTemp.X0.6.m | qfqm.tempStor.rateTemp.X0.7.m | qfqm.tempStor.rateTemp.X0.8.m | qfqm.tempStor.rateTemp.X0.9.m | qfqm.tempStor.rateTemp.X1.m | qfqm.tempStor.rateTemp.X1.1.m | qfqm.tempStor.rateTemp.X1.2.m | qfqm.tempStor.rateTemp.X1.3.m | qfqm.tempStor.rateTemp.X1.4.m | qfqm.tempStor.rateTemp.X1.5.m | qfqm.tempStor.rateTemp.X1.6.m | qfqm.tempStor.rateTemp.X1.7.m | qfqm.tempStor.rateTemp.X1.8.m | qfqm.tempStor.rateTemp.X1.9.m | qfqm.tempStor.rateTemp.X2.m | qfqm.tempStor.rateTemp.X2.1.m | qfqm.tempStor.rateTemp.X2.2.m | qfqm.tempStor.rateTemp.X2.3.m | qfqm.tempStor.rateTemp.X2.4.m | qfqm.tempStor.rateTemp.X2.5.m | qfqm.tempStor.rateTemp.X2.6.m | qfqm.tempStor.rateTemp.X2.7.m | qfqm.tempStor.rateTemp.X2.8.m | qfqm.tempStor.rateTemp.X2.9.m | qfqm.tempStor.rateTemp.X3.m | qfqm.tempStor.rateTemp.X3.1.m | qfqm.tempStor.rateTemp.X3.2.m | qfqm.tempStor.rateTemp.X3.3.m | qfqm.tempStor.rateTemp.X3.4.m | qfqm.tempStor.rateTemp.X3.5.m | qfqm.tempStor.rateTemp.X3.6.m | qfqm.tempStor.rateTemp.X3.7.m | qfqm.tempStor.rateTemp.X3.8.m | qfqm.tempStor.rateTemp.X3.9.m | qfqm.tempStor.rateTemp.X4.m | qfqm.tempStor.rateTemp.X4.1.m | qfqm.tempStor.rateTemp.X4.2.m | qfqm.tempStor.rateTemp.X4.3.m | qfqm.tempStor.rateTemp.X4.4.m | qfqm.tempStor.rateTemp.X4.5.m | qfqm.tempStor.rateTemp.X4.6.m | qfqm.tempStor.rateTemp.X4.7.m | qfqm.tempStor.rateTemp.X4.8.m | qfqm.tempStor.rateTemp.X4.9.m | qfqm.tempStor.rateTemp.X5.m | qfqm.tempStor.rateTemp.X5.1.m | qfqm.tempStor.rateTemp.X5.2.m | qfqm.tempStor.rateTemp.X5.3.m | qfqm.tempStor.rateTemp.X5.4.m | qfqm.tempStor.rateTemp.X5.5.m | qfqm.tempStor.rateTemp.X5.6.m | qfqm.tempStor.rateTemp.X5.7.m | qfqm.tempStor.rateTemp.X5.8.m | qfqm.tempStor.rateTemp.X5.9.m | qfqm.tempStor.rateTemp.X6.m | qfqm.tempStor.rateTemp.X6.1.m | qfqm.tempStor.rateTemp.X6.2.m | qfqm.tempStor.rateTemp.X6.3.m | qfqm.tempStor.rateTemp.X6.4.m | qfqm.tempStor.rateTemp.X6.5.m | qfqm.tempStor.rateTemp.X6.6.m | qfqm.tempStor.rateTemp.X6.7.m | qfqm.tempStor.rateTemp.X6.8.m | qfqm.tempStor.rateTemp.X6.9.m | qfqm.tempStor.rateTemp.X7.m | qfqm.tempStor.rateTemp.X7.1.m | qfqm.tempStor.rateTemp.X7.2.m | qfqm.tempStor.rateTemp.X7.3.m | qfqm.tempStor.rateTemp.X7.4.m | qfqm.tempStor.rateTemp.X7.5.m | qfqm.tempStor.rateTemp.X7.6.m | qfqm.tempStor.rateTemp.X7.7.m | qfqm.tempStor.rateTemp.X7.8.m | qfqm.tempStor.rateTemp.X7.9.m | qfqm.tempStor.rateTemp.X8.m | qfqm.tempStor.rateTemp.X8.1.m | qfqm.tempStor.rateTemp.X8.2.m | qfqm.tempStor.rateTemp.X8.3.m | qfqm.tempStor.rateTemp.X8.4.m |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2018-06-01 00:00:00 | 2018-06-01 00:29:59 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | -0.0002682 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | 0.0003159 | -0.0001014 | -0.0001014 | -0.0001014 | -0.0001014 | -0.0001014 | -0.0001051 | -0.0001112 | -0.0001172 | -0.0001233 | -0.0001294 | -0.0001354 | -0.0001415 | -0.0001476 | -0.0001537 | -0.0001597 | -0.0001658 | -0.0001719 | -0.0001779 | -0.0001840 | -0.0001857 | -0.0001870 | -0.0001882 | -0.0001895 | -0.0001907 | -0.0001919 | -0.0001932 | -0.0001944 | -0.0001956 | -0.0001969 | -0.0001981 | -0.0001994 | -0.0002006 | -0.0002018 | -0.0002031 | -0.0002043 | -0.0002055 | -0.0002068 | -0.0002080 | -0.0002093 | -0.0002105 | -0.0002117 | -0.0002130 | -0.0002142 | -0.0002154 | -0.0002172 | -0.0002190 | -0.0002208 | -0.0002225 | -0.0002243 | -0.0002261 | -0.0002278 | -0.0002296 | -0.0002314 | -0.0002332 | -0.0002349 | -0.0002367 | -0.0002385 | -0.0002402 | -0.0002420 | -0.0002438 | -0.0002456 | -0.0002473 | -0.0002491 | -0.0002509 | -0.0002526 | -0.0002544 | -0.0002562 | -0.0002580 | -0.0002597 | -0.0002615 | -0.0002633 | -0.0002651 | -0.0002668 | -0.0002686 | -0.0002704 | -0.0002721 | -0.0002739 | -0.0002757 | -0.0002775 | -0.0002792 | -0.0002810 | -0.0002828 | -0.0002845 | -0.0002863 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2018-06-01 00:30:00 | 2018-06-01 00:59:59 | 0.0004879 | 0.0004879 | 0.0004879 | 0.0004879 | 0.0004879 | 0.0004674 | 0.0004331 | 0.0003989 | 0.0003647 | 0.0003305 | 0.0002963 | 0.0002621 | 0.0002278 | 0.0001936 | 0.0001594 | 0.0001252 | 0.0000910 | 0.0000568 | 0.0000225 | 0.0000381 | 0.0000592 | 0.0000803 | 0.0001013 | 0.0001224 | 0.0001435 | 0.0001646 | 0.0001857 | 0.0002068 | 0.0002278 | 0.0002489 | 0.0002700 | 0.0002911 | 0.0003122 | 0.0003333 | 0.0003543 | 0.0003754 | 0.0003965 | 0.0004176 | 0.0004387 | 0.0004598 | 0.0004808 | 0.0005019 | 0.0005230 | 0.0005441 | 0.0005393 | 0.0005346 | 0.0005298 | 0.0005251 | 0.0005203 | 0.0005156 | 0.0005108 | 0.0005061 | 0.0005013 | 0.0004966 | 0.0004918 | 0.0004871 | 0.0004823 | 0.0004776 | 0.0004728 | 0.0004681 | 0.0004633 | 0.0004586 | 0.0004538 | 0.0004491 | 0.0004443 | 0.0004396 | 0.0004348 | 0.0004301 | 0.0004253 | 0.0004206 | 0.0004158 | 0.0004111 | 0.0004063 | 0.0004016 | 0.0003968 | 0.0003921 | 0.0003873 | 0.0003826 | 0.0003778 | 0.0003731 | 0.0003683 | 0.0003636 | 0.0003588 | 0.0003541 | -0.0003565 | -0.0003565 | -0.0003565 | -0.0003565 | -0.0003565 | -0.0003426 | -0.0003192 | -0.0002959 | -0.0002726 | -0.0002493 | -0.0002260 | -0.0002027 | -0.0001793 | -0.0001560 | -0.0001327 | -0.0001094 | -0.0000861 | -0.0000628 | -0.0000394 | -0.0000366 | -0.0000361 | -0.0000356 | -0.0000350 | -0.0000345 | -0.0000339 | -0.0000334 | -0.0000329 | -0.0000323 | -0.0000318 | -0.0000313 | -0.0000307 | -0.0000302 | -0.0000296 | -0.0000291 | -0.0000286 | -0.0000280 | -0.0000275 | -0.0000270 | -0.0000264 | -0.0000259 | -0.0000253 | -0.0000248 | -0.0000243 | -0.0000237 | -0.0000279 | -0.0000321 | -0.0000363 | -0.0000405 | -0.0000446 | -0.0000488 | -0.0000530 | -0.0000572 | -0.0000614 | -0.0000656 | -0.0000697 | -0.0000739 | -0.0000781 | -0.0000823 | -0.0000865 | -0.0000906 | -0.0000948 | -0.0000990 | -0.0001032 | -0.0001074 | -0.0001116 | -0.0001157 | -0.0001199 | -0.0001241 | -0.0001283 | -0.0001325 | -0.0001366 | -0.0001408 | -0.0001450 | -0.0001492 | -0.0001534 | -0.0001576 | -0.0001617 | -0.0001659 | -0.0001701 | -0.0001743 | -0.0001785 | -0.0001827 | -0.0001868 | -0.0001910 | -0.0001814 | -0.0001814 | -0.0001814 | -0.0001814 | -0.0001814 | -0.0001725 | -0.0001576 | -0.0001427 | -0.0001278 | -0.0001129 | -0.0000980 | -0.0000831 | -0.0000683 | -0.0000534 | -0.0000385 | -0.0000236 | -0.0000087 | 0.0000062 | 0.0000211 | 0.0000247 | 0.0000271 | 0.0000295 | 0.0000318 | 0.0000342 | 0.0000366 | 0.0000390 | 0.0000414 | 0.0000437 | 0.0000461 | 0.0000485 | 0.0000509 | 0.0000533 | 0.0000556 | 0.0000580 | 0.0000604 | 0.0000628 | 0.0000652 | 0.0000675 | 0.0000699 | 0.0000723 | 0.0000747 | 0.0000771 | 0.0000794 | 0.0000818 | 0.0000836 | 0.0000854 | 0.0000872 | 0.0000890 | 0.0000908 | 0.0000926 | 0.0000944 | 0.0000962 | 0.0000980 | 0.0000998 | 0.0001016 | 0.0001033 | 0.0001051 | 0.0001069 | 0.0001087 | 0.0001105 | 0.0001123 | 0.0001141 | 0.0001159 | 0.0001177 | 0.0001195 | 0.0001213 | 0.0001231 | 0.0001249 | 0.0001267 | 0.0001285 | 0.0001303 | 0.0001320 | 0.0001338 | 0.0001356 | 0.0001374 | 0.0001392 | 0.0001410 | 0.0001428 | 0.0001446 | 0.0001464 | 0.0001482 | 0.0001500 | 0.0001518 | 0.0001536 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2018-06-01 01:00:00 | 2018-06-01 01:29:59 | 0.0005086 | 0.0005086 | 0.0005086 | 0.0005086 | 0.0005086 | 0.0005025 | 0.0004925 | 0.0004825 | 0.0004724 | 0.0004624 | 0.0004523 | 0.0004423 | 0.0004323 | 0.0004222 | 0.0004122 | 0.0004021 | 0.0003921 | 0.0003820 | 0.0003720 | 0.0003480 | 0.0003224 | 0.0002968 | 0.0002712 | 0.0002457 | 0.0002201 | 0.0001945 | 0.0001689 | 0.0001434 | 0.0001178 | 0.0000922 | 0.0000666 | 0.0000410 | 0.0000155 | -0.0000101 | -0.0000357 | -0.0000613 | -0.0000869 | -0.0001124 | -0.0001380 | -0.0001636 | -0.0001892 | -0.0002147 | -0.0002403 | -0.0002659 | -0.0002551 | -0.0002443 | -0.0002335 | -0.0002227 | -0.0002119 | -0.0002011 | -0.0001904 | -0.0001796 | -0.0001688 | -0.0001580 | -0.0001472 | -0.0001364 | -0.0001256 | -0.0001148 | -0.0001040 | -0.0000932 | -0.0000824 | -0.0000716 | -0.0000609 | -0.0000501 | -0.0000393 | -0.0000285 | -0.0000177 | -0.0000069 | 0.0000039 | 0.0000147 | 0.0000255 | 0.0000363 | 0.0000471 | 0.0000579 | 0.0000686 | 0.0000794 | 0.0000902 | 0.0001010 | 0.0001118 | 0.0001226 | 0.0001334 | 0.0001442 | 0.0001550 | 0.0001658 | -0.0002073 | -0.0002073 | -0.0002073 | -0.0002073 | -0.0002073 | -0.0002152 | -0.0002283 | -0.0002414 | -0.0002545 | -0.0002676 | -0.0002807 | -0.0002938 | -0.0003069 | -0.0003200 | -0.0003331 | -0.0003462 | -0.0003593 | -0.0003724 | -0.0003855 | -0.0003947 | -0.0004034 | -0.0004122 | -0.0004209 | -0.0004297 | -0.0004384 | -0.0004472 | -0.0004559 | -0.0004647 | -0.0004735 | -0.0004822 | -0.0004910 | -0.0004997 | -0.0005085 | -0.0005172 | -0.0005260 | -0.0005347 | -0.0005435 | -0.0005523 | -0.0005610 | -0.0005698 | -0.0005785 | -0.0005873 | -0.0005960 | -0.0006048 | -0.0005965 | -0.0005881 | -0.0005798 | -0.0005714 | -0.0005631 | -0.0005548 | -0.0005464 | -0.0005381 | -0.0005297 | -0.0005214 | -0.0005131 | -0.0005047 | -0.0004964 | -0.0004880 | -0.0004797 | -0.0004713 | -0.0004630 | -0.0004547 | -0.0004463 | -0.0004380 | -0.0004296 | -0.0004213 | -0.0004130 | -0.0004046 | -0.0003963 | -0.0003879 | -0.0003796 | -0.0003713 | -0.0003629 | -0.0003546 | -0.0003462 | -0.0003379 | -0.0003296 | -0.0003212 | -0.0003129 | -0.0003045 | -0.0002962 | -0.0002879 | -0.0002795 | -0.0002712 | -0.0002556 | -0.0002556 | -0.0002556 | -0.0002556 | -0.0002556 | -0.0002458 | -0.0002293 | -0.0002129 | -0.0001965 | -0.0001801 | -0.0001637 | -0.0001473 | -0.0001308 | -0.0001144 | -0.0000980 | -0.0000816 | -0.0000652 | -0.0000487 | -0.0000323 | -0.0000270 | -0.0000229 | -0.0000188 | -0.0000147 | -0.0000106 | -0.0000065 | -0.0000024 | 0.0000017 | 0.0000058 | 0.0000099 | 0.0000140 | 0.0000181 | 0.0000222 | 0.0000263 | 0.0000304 | 0.0000345 | 0.0000386 | 0.0000427 | 0.0000468 | 0.0000509 | 0.0000550 | 0.0000591 | 0.0000632 | 0.0000673 | 0.0000714 | 0.0000739 | 0.0000765 | 0.0000790 | 0.0000815 | 0.0000840 | 0.0000866 | 0.0000891 | 0.0000916 | 0.0000941 | 0.0000967 | 0.0000992 | 0.0001017 | 0.0001042 | 0.0001068 | 0.0001093 | 0.0001118 | 0.0001143 | 0.0001169 | 0.0001194 | 0.0001219 | 0.0001244 | 0.0001270 | 0.0001295 | 0.0001320 | 0.0001345 | 0.0001371 | 0.0001396 | 0.0001421 | 0.0001446 | 0.0001472 | 0.0001497 | 0.0001522 | 0.0001547 | 0.0001573 | 0.0001598 | 0.0001623 | 0.0001648 | 0.0001674 | 0.0001699 | 0.0001724 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2018-06-01 01:30:00 | 2018-06-01 01:59:59 | 0.0013277 | 0.0013277 | 0.0013277 | 0.0013277 | 0.0013277 | 0.0013735 | 0.0014499 | 0.0015263 | 0.0016027 | 0.0016790 | 0.0017554 | 0.0018318 | 0.0019082 | 0.0019845 | 0.0020609 | 0.0021373 | 0.0022137 | 0.0022900 | 0.0023664 | 0.0023013 | 0.0022204 | 0.0021396 | 0.0020587 | 0.0019778 | 0.0018970 | 0.0018161 | 0.0017353 | 0.0016544 | 0.0015735 | 0.0014927 | 0.0014118 | 0.0013310 | 0.0012501 | 0.0011692 | 0.0010884 | 0.0010075 | 0.0009267 | 0.0008458 | 0.0007649 | 0.0006841 | 0.0006032 | 0.0005224 | 0.0004415 | 0.0003606 | 0.0003730 | 0.0003853 | 0.0003976 | 0.0004099 | 0.0004222 | 0.0004345 | 0.0004468 | 0.0004591 | 0.0004714 | 0.0004837 | 0.0004960 | 0.0005083 | 0.0005206 | 0.0005329 | 0.0005452 | 0.0005575 | 0.0005698 | 0.0005821 | 0.0005944 | 0.0006067 | 0.0006190 | 0.0006313 | 0.0006436 | 0.0006559 | 0.0006682 | 0.0006805 | 0.0006928 | 0.0007051 | 0.0007174 | 0.0007297 | 0.0007420 | 0.0007543 | 0.0007666 | 0.0007789 | 0.0007912 | 0.0008035 | 0.0008158 | 0.0008281 | 0.0008404 | 0.0008527 | 0.0000313 | 0.0000313 | 0.0000313 | 0.0000313 | 0.0000313 | 0.0000337 | 0.0000378 | 0.0000418 | 0.0000459 | 0.0000499 | 0.0000540 | 0.0000580 | 0.0000620 | 0.0000661 | 0.0000701 | 0.0000742 | 0.0000782 | 0.0000823 | 0.0000863 | 0.0000849 | 0.0000830 | 0.0000810 | 0.0000791 | 0.0000771 | 0.0000751 | 0.0000732 | 0.0000712 | 0.0000692 | 0.0000673 | 0.0000653 | 0.0000634 | 0.0000614 | 0.0000594 | 0.0000575 | 0.0000555 | 0.0000536 | 0.0000516 | 0.0000496 | 0.0000477 | 0.0000457 | 0.0000437 | 0.0000418 | 0.0000398 | 0.0000379 | 0.0000349 | 0.0000319 | 0.0000290 | 0.0000260 | 0.0000230 | 0.0000201 | 0.0000171 | 0.0000141 | 0.0000112 | 0.0000082 | 0.0000052 | 0.0000023 | -0.0000007 | -0.0000037 | -0.0000066 | -0.0000096 | -0.0000126 | -0.0000155 | -0.0000185 | -0.0000215 | -0.0000244 | -0.0000274 | -0.0000304 | -0.0000333 | -0.0000363 | -0.0000393 | -0.0000422 | -0.0000452 | -0.0000482 | -0.0000511 | -0.0000541 | -0.0000571 | -0.0000600 | -0.0000630 | -0.0000660 | -0.0000689 | -0.0000719 | -0.0000749 | -0.0000778 | -0.0000808 | -0.0010983 | -0.0010983 | -0.0010983 | -0.0010983 | -0.0010983 | -0.0010630 | -0.0010043 | -0.0009456 | -0.0008868 | -0.0008281 | -0.0007693 | -0.0007106 | -0.0006519 | -0.0005931 | -0.0005344 | -0.0004756 | -0.0004169 | -0.0003582 | -0.0002994 | -0.0002907 | -0.0002876 | -0.0002845 | -0.0002814 | -0.0002783 | -0.0002751 | -0.0002720 | -0.0002689 | -0.0002658 | -0.0002627 | -0.0002595 | -0.0002564 | -0.0002533 | -0.0002502 | -0.0002471 | -0.0002439 | -0.0002408 | -0.0002377 | -0.0002346 | -0.0002315 | -0.0002283 | -0.0002252 | -0.0002221 | -0.0002190 | -0.0002159 | -0.0002160 | -0.0002161 | -0.0002162 | -0.0002163 | -0.0002164 | -0.0002165 | -0.0002166 | -0.0002167 | -0.0002168 | -0.0002169 | -0.0002170 | -0.0002171 | -0.0002173 | -0.0002174 | -0.0002175 | -0.0002176 | -0.0002177 | -0.0002178 | -0.0002179 | -0.0002180 | -0.0002181 | -0.0002182 | -0.0002183 | -0.0002184 | -0.0002185 | -0.0002186 | -0.0002188 | -0.0002189 | -0.0002190 | -0.0002191 | -0.0002192 | -0.0002193 | -0.0002194 | -0.0002195 | -0.0002196 | -0.0002197 | -0.0002198 | -0.0002199 | -0.0002200 | -0.0002201 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2018-06-01 02:00:00 | 2018-06-01 02:29:59 | 0.0007344 | 0.0007344 | 0.0007344 | 0.0007344 | 0.0007344 | 0.0008510 | 0.0010454 | 0.0012397 | 0.0014341 | 0.0016284 | 0.0018228 | 0.0020171 | 0.0022115 | 0.0024058 | 0.0026002 | 0.0027946 | 0.0029889 | 0.0031833 | 0.0033776 | 0.0032919 | 0.0031751 | 0.0030583 | 0.0029415 | 0.0028247 | 0.0027079 | 0.0025911 | 0.0024743 | 0.0023575 | 0.0022407 | 0.0021239 | 0.0020071 | 0.0018903 | 0.0017735 | 0.0016567 | 0.0015399 | 0.0014231 | 0.0013063 | 0.0011895 | 0.0010727 | 0.0009558 | 0.0008390 | 0.0007222 | 0.0006054 | 0.0004886 | 0.0004664 | 0.0004442 | 0.0004219 | 0.0003997 | 0.0003775 | 0.0003553 | 0.0003330 | 0.0003108 | 0.0002886 | 0.0002664 | 0.0002441 | 0.0002219 | 0.0001997 | 0.0001774 | 0.0001552 | 0.0001330 | 0.0001108 | 0.0000885 | 0.0000663 | 0.0000441 | 0.0000218 | -0.0000004 | -0.0000226 | -0.0000448 | -0.0000671 | -0.0000893 | -0.0001115 | -0.0001338 | -0.0001560 | -0.0001782 | -0.0002004 | -0.0002227 | -0.0002449 | -0.0002671 | -0.0002894 | -0.0003116 | -0.0003338 | -0.0003560 | -0.0003783 | -0.0004005 | -0.0000316 | -0.0000316 | -0.0000316 | -0.0000316 | -0.0000316 | -0.0000246 | -0.0000130 | -0.0000013 | 0.0000103 | 0.0000219 | 0.0000336 | 0.0000452 | 0.0000568 | 0.0000685 | 0.0000801 | 0.0000917 | 0.0001034 | 0.0001150 | 0.0001266 | 0.0001239 | 0.0001195 | 0.0001152 | 0.0001109 | 0.0001065 | 0.0001022 | 0.0000978 | 0.0000935 | 0.0000891 | 0.0000848 | 0.0000804 | 0.0000761 | 0.0000717 | 0.0000674 | 0.0000630 | 0.0000587 | 0.0000543 | 0.0000500 | 0.0000456 | 0.0000413 | 0.0000369 | 0.0000326 | 0.0000283 | 0.0000239 | 0.0000196 | 0.0000170 | 0.0000145 | 0.0000120 | 0.0000095 | 0.0000070 | 0.0000045 | 0.0000020 | -0.0000005 | -0.0000030 | -0.0000056 | -0.0000081 | -0.0000106 | -0.0000131 | -0.0000156 | -0.0000181 | -0.0000206 | -0.0000231 | -0.0000256 | -0.0000282 | -0.0000307 | -0.0000332 | -0.0000357 | -0.0000382 | -0.0000407 | -0.0000432 | -0.0000457 | -0.0000482 | -0.0000508 | -0.0000533 | -0.0000558 | -0.0000583 | -0.0000608 | -0.0000633 | -0.0000658 | -0.0000683 | -0.0000708 | -0.0000733 | -0.0000759 | -0.0000784 | -0.0000809 | -0.0006432 | -0.0006432 | -0.0006432 | -0.0006432 | -0.0006432 | -0.0006189 | -0.0005784 | -0.0005379 | -0.0004973 | -0.0004568 | -0.0004163 | -0.0003758 | -0.0003353 | -0.0002947 | -0.0002542 | -0.0002137 | -0.0001732 | -0.0001326 | -0.0000921 | -0.0000846 | -0.0000808 | -0.0000770 | -0.0000732 | -0.0000694 | -0.0000656 | -0.0000618 | -0.0000580 | -0.0000541 | -0.0000503 | -0.0000465 | -0.0000427 | -0.0000389 | -0.0000351 | -0.0000313 | -0.0000275 | -0.0000236 | -0.0000198 | -0.0000160 | -0.0000122 | -0.0000084 | -0.0000046 | -0.0000008 | 0.0000030 | 0.0000069 | 0.0000066 | 0.0000064 | 0.0000062 | 0.0000059 | 0.0000057 | 0.0000055 | 0.0000052 | 0.0000050 | 0.0000048 | 0.0000045 | 0.0000043 | 0.0000041 | 0.0000039 | 0.0000036 | 0.0000034 | 0.0000032 | 0.0000029 | 0.0000027 | 0.0000025 | 0.0000022 | 0.0000020 | 0.0000018 | 0.0000015 | 0.0000013 | 0.0000011 | 0.0000008 | 0.0000006 | 0.0000004 | 0.0000002 | -0.0000001 | -0.0000003 | -0.0000005 | -0.0000008 | -0.0000010 | -0.0000012 | -0.0000015 | -0.0000017 | -0.0000019 | -0.0000022 | -0.0000024 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2018-06-01 02:30:00 | 2018-06-01 02:59:59 | -0.0009450 | -0.0009450 | -0.0009450 | -0.0009450 | -0.0009450 | -0.0007653 | -0.0004659 | -0.0001665 | 0.0001329 | 0.0004323 | 0.0007317 | 0.0010311 | 0.0013305 | 0.0016300 | 0.0019294 | 0.0022288 | 0.0025282 | 0.0028276 | 0.0031270 | 0.0030239 | 0.0028760 | 0.0027282 | 0.0025803 | 0.0024325 | 0.0022846 | 0.0021368 | 0.0019889 | 0.0018411 | 0.0016932 | 0.0015454 | 0.0013975 | 0.0012497 | 0.0011018 | 0.0009540 | 0.0008061 | 0.0006583 | 0.0005104 | 0.0003626 | 0.0002147 | 0.0000669 | -0.0000810 | -0.0002288 | -0.0003767 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0005245 | -0.0001513 | -0.0001513 | -0.0001513 | -0.0001513 | -0.0001513 | -0.0001414 | -0.0001250 | -0.0001085 | -0.0000921 | -0.0000756 | -0.0000591 | -0.0000427 | -0.0000262 | -0.0000098 | 0.0000067 | 0.0000232 | 0.0000396 | 0.0000561 | 0.0000725 | 0.0000696 | 0.0000645 | 0.0000594 | 0.0000544 | 0.0000493 | 0.0000442 | 0.0000391 | 0.0000340 | 0.0000289 | 0.0000238 | 0.0000188 | 0.0000137 | 0.0000086 | 0.0000035 | -0.0000016 | -0.0000067 | -0.0000118 | -0.0000168 | -0.0000219 | -0.0000270 | -0.0000321 | -0.0000372 | -0.0000423 | -0.0000474 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0000524 | -0.0010168 | -0.0010168 | -0.0010168 | -0.0010168 | -0.0010168 | -0.0009788 | -0.0009156 | -0.0008524 | -0.0007891 | -0.0007259 | -0.0006627 | -0.0005994 | -0.0005362 | -0.0004729 | -0.0004097 | -0.0003465 | -0.0002832 | -0.0002200 | -0.0001568 | -0.0001510 | -0.0001516 | -0.0001523 | -0.0001529 | -0.0001535 | -0.0001542 | -0.0001548 | -0.0001554 | -0.0001561 | -0.0001567 | -0.0001574 | -0.0001580 | -0.0001586 | -0.0001593 | -0.0001599 | -0.0001605 | -0.0001612 | -0.0001618 | -0.0001624 | -0.0001631 | -0.0001637 | -0.0001643 | -0.0001650 | -0.0001656 | -0.0001662 | -0.0001655 | -0.0001647 | -0.0001639 | -0.0001631 | -0.0001623 | -0.0001615 | -0.0001607 | -0.0001600 | -0.0001592 | -0.0001584 | -0.0001576 | -0.0001568 | -0.0001560 | -0.0001552 | -0.0001545 | -0.0001537 | -0.0001529 | -0.0001521 | -0.0001513 | -0.0001505 | -0.0001497 | -0.0001490 | -0.0001482 | -0.0001474 | -0.0001466 | -0.0001458 | -0.0001450 | -0.0001442 | -0.0001435 | -0.0001427 | -0.0001419 | -0.0001411 | -0.0001403 | -0.0001395 | -0.0001387 | -0.0001380 | -0.0001372 | -0.0001364 | -0.0001356 | -0.0001348 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5.6.7 Un-interpolated vertical profile data (Level 2)
The Level 2 data are interpolated in time but not in space. They contain the rates of change at the measurement heights.
Again, they can be extracted from the HDF5 files using stackEddy()
with the same syntax:
prof.l2 <- stackEddy(filepath="./data/filesToStack00200/",
level="dp02")## Extracting data
##
|
| | 0%
|
|================== | 25%
|
|=================================== | 50%
|
|==================================================== | 75%
|
|======================================================================| 100%
## Stacking data tables by month
##
|
| | 0%
|
|= | 2%
|
|== | 3%
|
|==== | 5%
|
|===== | 7%
|
|====== | 8%
|
|======= | 10%
|
|======== | 12%
|
|========= | 13%
|
|========== | 15%
|
|============ | 17%
|
|============= | 18%
|
|============== | 20%
|
|=============== | 22%
|
|================ | 23%
|
|================== | 25%
|
|=================== | 27%
|
|==================== | 28%
|
|===================== | 30%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 35%
|
|========================== | 37%
|
|=========================== | 38%
|
|============================ | 40%
|
|============================= | 42%
|
|============================== | 43%
|
|================================ | 45%
|
|================================= | 47%
|
|================================== | 48%
|
|=================================== | 50%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 55%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================== | 60%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================== | 65%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================= | 70%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 75%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================== | 80%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|============================================================ | 85%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|=============================================================== | 90%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 95%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|======================================================================| 100%
## Joining data variables
##
|
| | 0%
|
|====== | 8%
|
|============ | 17%
|
|================== | 25%
|
|======================= | 33%
|
|============================= | 42%
|
|=================================== | 50%
|
|========================================= | 58%
|
|=============================================== | 67%
|
|==================================================== | 75%
|
|========================================================== | 83%
|
|================================================================ | 92%
|
|======================================================================| 100%knitr::kable(head(prof.l2$HARV))Note that here, as in the PAR data, there is a verticalPosition field.
It has the same meaning as in the PAR data, indicating the tower level
of the measurement.
5.6.8 Calibrated raw data (Level 1)
Level 1 (dp01) data are calibrated, and aggregated in time, but
otherwise untransformed. Use Level 1 data for raw gas concentrations and
atmospheric stable isotopes.
Using stackEddy() to extract Level 1 data requires additional inputs.
The Level 1 files are too large to simply pull out all the variables by
default, and they include mutiple averaging intervals, which can’t be
merged. So two additional inputs are needed:
avg: The averaging interval to extractvar: One or more variables to extract
What variables are available, at what averaging intervals? Another
function in the neonUtilities package, getVarsEddy(), returns a list
of HDF5 file contents. It requires only one input, a filepath to a
single NEON HDF5 file:
files <- list.files("./data/filesToStack00200")
vars <- getVarsEddy(paste0("./data/filesToStack00200/",
files[grep(pattern = paste0("HARV", ".*.", ".*.h5$"),
x = files)][1]))
knitr::kable(head(vars))| site | level | category | system | hor | ver | tmi | subsys | name | otype | dclass | dim | oth | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | HARV | dp01 | data | amrs | 000 | 060 | 01m | NA | angNedXaxs | H5I_DATASET | COMPOUND | 43200 | NA |
| 6 | HARV | dp01 | data | amrs | 000 | 060 | 01m | NA | angNedYaxs | H5I_DATASET | COMPOUND | 43200 | NA |
| 7 | HARV | dp01 | data | amrs | 000 | 060 | 01m | NA | angNedZaxs | H5I_DATASET | COMPOUND | 43200 | NA |
| 9 | HARV | dp01 | data | amrs | 000 | 060 | 30m | NA | angNedXaxs | H5I_DATASET | COMPOUND | 1440 | NA |
| 10 | HARV | dp01 | data | amrs | 000 | 060 | 30m | NA | angNedYaxs | H5I_DATASET | COMPOUND | 1440 | NA |
| 11 | HARV | dp01 | data | amrs | 000 | 060 | 30m | NA | angNedZaxs | H5I_DATASET | COMPOUND | 1440 | NA |
Inputs to var can be any values from the name field in the table
returned by getVarsEddy(). Let’s take a look at CO2 and
H2O, 13C in CO2 and 18O in
H2O, at 30-minute aggregation. Let’s look at Harvard Forest
for these data, since deeper canopies generally have more interesting
profiles:
iso <- stackEddy(filepath="./data/filesToStack00200/",
level="dp01", var=c("rtioMoleDryCo2","rtioMoleDryH2o",
"dlta13CCo2","dlta18OH2o"), avg=30)## Extracting data
##
|
| | 0%
|
|================== | 25%
|
|=================================== | 50%
|
|==================================================== | 75%
|
|======================================================================| 100%
## Stacking data tables by month
##
|
| | 0%
|
| | 1%
|
|= | 1%
|
|= | 2%
|
|== | 2%
|
|== | 3%
|
|=== | 4%
|
|=== | 5%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 8%
|
|====== | 9%
|
|======= | 9%
|
|======= | 10%
|
|======= | 11%
|
|======== | 11%
|
|======== | 12%
|
|========= | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============ | 18%
|
|============= | 18%
|
|============= | 19%
|
|============== | 19%
|
|============== | 20%
|
|============== | 21%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 22%
|
|================ | 23%
|
|================= | 24%
|
|================= | 25%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|=================== | 28%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 29%
|
|===================== | 30%
|
|===================== | 31%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================ | 41%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|=============================== | 45%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================= | 48%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 49%
|
|=================================== | 50%
|
|=================================== | 51%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 59%
|
|========================================== | 60%
|
|========================================== | 61%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|=============================================== | 68%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 69%
|
|================================================= | 70%
|
|================================================= | 71%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|====================================================== | 78%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 79%
|
|======================================================== | 80%
|
|======================================================== | 81%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================= | 88%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 89%
|
|=============================================================== | 90%
|
|=============================================================== | 91%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|==================================================================== | 98%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 99%
|
|======================================================================| 100%
## Joining data variables
##
|
| | 0%
|
|= | 2%
|
|=== | 4%
|
|==== | 6%
|
|===== | 7%
|
|====== | 9%
|
|======== | 11%
|
|========= | 13%
|
|========== | 15%
|
|============ | 17%
|
|============= | 19%
|
|============== | 20%
|
|================ | 22%
|
|================= | 24%
|
|================== | 26%
|
|=================== | 28%
|
|===================== | 30%
|
|====================== | 31%
|
|======================= | 33%
|
|========================= | 35%
|
|========================== | 37%
|
|=========================== | 39%
|
|============================= | 41%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 46%
|
|================================== | 48%
|
|=================================== | 50%
|
|==================================== | 52%
|
|====================================== | 54%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 59%
|
|=========================================== | 61%
|
|============================================ | 63%
|
|============================================= | 65%
|
|=============================================== | 67%
|
|================================================ | 69%
|
|================================================= | 70%
|
|=================================================== | 72%
|
|==================================================== | 74%
|
|===================================================== | 76%
|
|====================================================== | 78%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================== | 83%
|
|============================================================ | 85%
|
|============================================================= | 87%
|
|============================================================== | 89%
|
|================================================================ | 91%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|=================================================================== | 96%
|
|===================================================================== | 98%
|
|======================================================================| 100%knitr::kable(head(iso$HARV))Let’s plot vertical profiles of CO2 and 13C in CO2 on a single day.
Here, for convenience, instead of converting the time stamps to a time
format, it’s easy to use the character format to extract the ones we
want using grep(). And discard the verticalPosition values that are
string values - those are the calibration gases.
iso.d <- iso$HARV[grep("2018-06-25", iso$HARV$timeBgn, fixed=T),]
iso.d <- iso.d[-which(is.na(as.numeric(iso.d$verticalPosition))),]## Warning in which(is.na(as.numeric(iso.d$verticalPosition))): NAs introduced by
## coercionggplot is well suited to these types of data, let’s use it to plot the
profiles.
library(ggplot2)g <- ggplot(iso.d, aes(y=verticalPosition)) +
geom_path(aes(x=data.co2Stor.rtioMoleDryCo2.mean,
group=timeBgn, col=timeBgn)) +
theme(legend.position="none") +
xlab("CO2") + ylab("Tower level")
g## Warning: Removed 2 row(s) containing missing values (geom_path).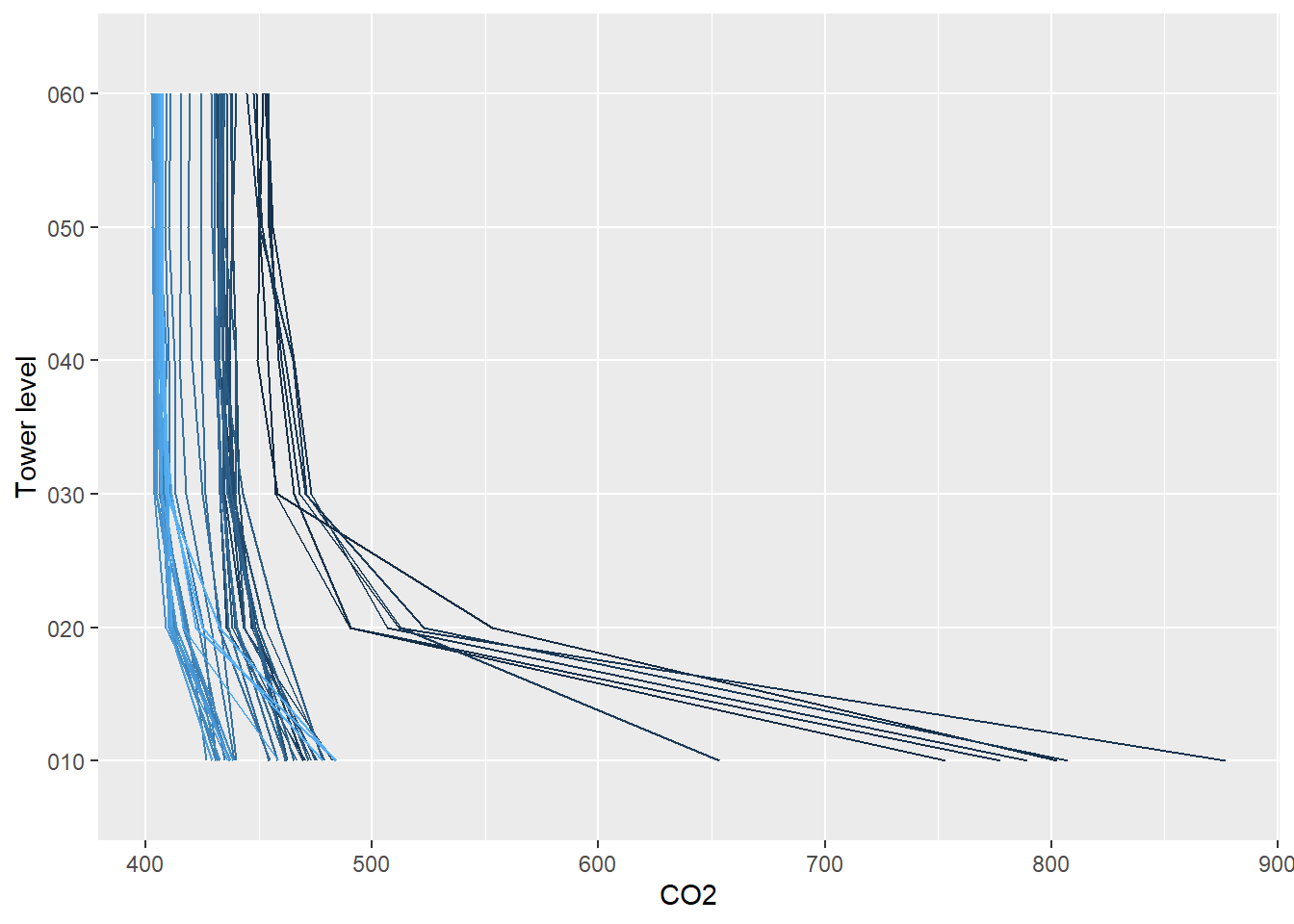
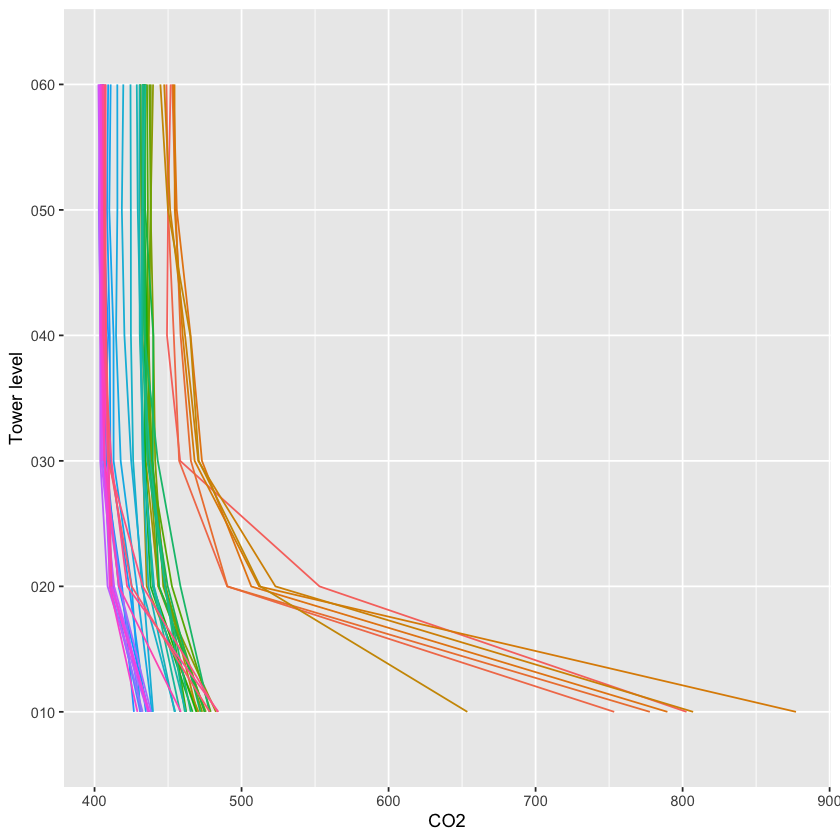
g <- ggplot(iso.d, aes(y=verticalPosition)) +
geom_path(aes(x=data.isoCo2.dlta13CCo2.mean,
group=timeBgn, col=timeBgn)) +
theme(legend.position="none") +
xlab("d13C") + ylab("Tower level")
g## Warning: Removed 55 row(s) containing missing values (geom_path).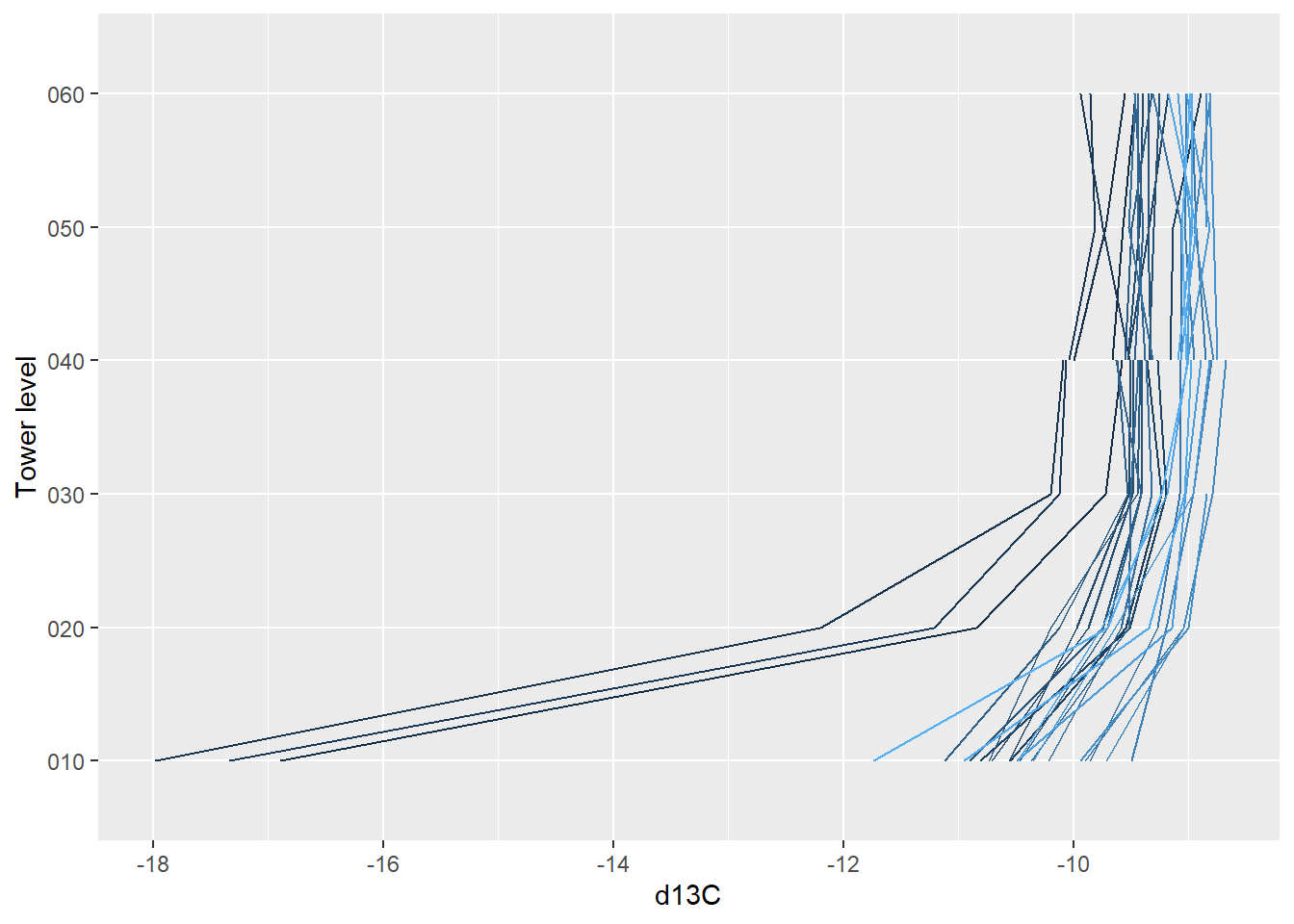
Warning message:
“Removed 55 rows containing missing values (geom_path).”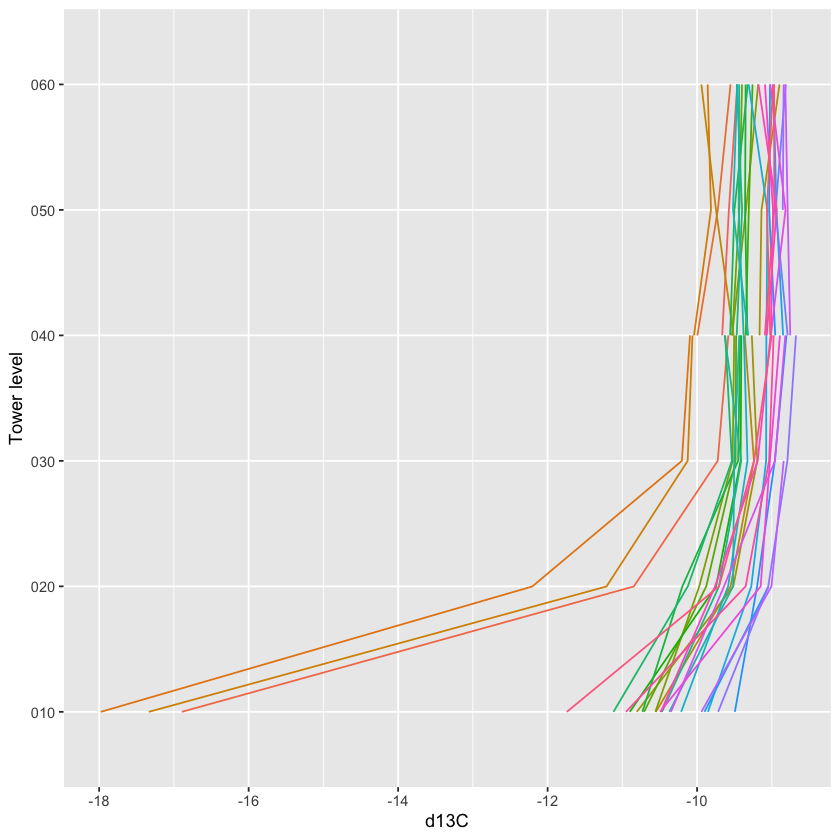
The legends are omitted for space, see if you can work out the times of day the different colors represent.
5.6.9 Convert NEON flux data variables to AmeriFlux FP standard
Install and load packages
#Install most recent version of ffbase from GitHub, this is a package dependency of eddy4R.base
devtools::install_github("edwindj/ffbase", subdir="pkg")## Skipping install of 'ffbase' from a github remote, the SHA1 (14f75881) has not changed since last install.
## Use `force = TRUE` to force installation#Install NEONprocIS.base from GitHub, this package is a dependency of eddy4R.base
devtools::install_github(repo="NEONScience/NEON-IS-data-processing",
ref="master",
subdir="pack/NEONprocIS.base",
dependencies=c(NA, TRUE)[2],
repos=c(BiocManager::repositories(), # for dependencies on Bioconductor packages
"https://cran.rstudio.com/") # for CRAN
)## Skipping install of 'NEONprocIS.base' from a github remote, the SHA1 (a98dafd9) has not changed since last install.
## Use `force = TRUE` to force installation#Install eddy4R.base from GitHub
devtools::install_github(repo="NEONScience/eddy4R",
ref="master",
subdir="pack/eddy4R.base",
dependencies=c(NA, TRUE)[2],
repos=c(BiocManager::repositories(), # for dependencies on Bioconductor packages
"https://cran.rstudio.com/") # for CRAN
)## Skipping install of 'eddy4R.base' from a github remote, the SHA1 (d77bd677) has not changed since last install.
## Use `force = TRUE` to force installationpackReq <- c("rhdf5", "eddy4R.base", "jsonlite", "lubridate")
lapply(packReq, function(x) {
print(x)
if(require(x, character.only = TRUE) == FALSE) {
install.packages(x)
library(x, character.only = TRUE)
}})## [1] "rhdf5"## Loading required package: rhdf5## [1] "eddy4R.base"## Loading required package: eddy4R.base## [1] "jsonlite"## Loading required package: jsonlite## [1] "lubridate"## Loading required package: lubridate## Warning: package 'lubridate' was built under R version 4.2.2## Loading required package: timechange## Warning: package 'timechange' was built under R version 4.2.2##
## Attaching package: 'lubridate'## The following objects are masked from 'package:base':
##
## date, intersect, setdiff, union## [[1]]
## NULL
##
## [[2]]
## NULL
##
## [[3]]
## NULL
##
## [[4]]
## NULLh5disableFileLocking()Select your site of interest from the list of NEON sites below.
site <- "KONZ"
#"BARR","CLBJ","MLBS","DSNY","NIWO","ORNL","OSBS",
#"SCBI","LENO","TALL","CPER","BART","HARV","BLAN",
#"SERC","JERC","GUAN","LAJA","STEI","TREE","UNDE",
#"KONA","KONZ","UKFS","GRSM","DELA","DCFS","NOGP",
#"WOOD","RMNP","OAES","YELL","MOAB","STER","JORN",
#"SRER","ONAQ","ABBY","WREF","SJER","SOAP","TEAK",
#"TOOL","BONA","DEJU","HEAL","PUUM"If you would like to download a set range of dates, define the following paramemters. If these are not defined, it will default to the entire record at the site
#define start and end dates, optional, defaults to entire period of site operation. Use %Y-%m-%d format.
dateBgn <- "2020-03-01"
dateEnd <- "2020-05-31"
# Data package from the portal
Pack <- c('basic','expanded')[1]
#The version data for the FP standard conversion processing
ver = paste0("v",format(Sys.time(), "%Y%m%dT%H%m"))Specify Download directory for HDF5 files from the NEON data portal and output directory to save the resulting csv files. Change save paths to where you want the files on your computer.
#download directory
DirDnld=tempdir()
#Output directory, change this to where you want to save the output csv
DirOutBase <-paste0("~/eddy/data/Ameriflux/",ver)Specify Data Product number, for the Bundled Eddy-Covariance files, this is DP4.00200.001
#DP number
dpID <- 'DP4.00200.001'Get metadata from Ameriflux Site Info BADM sheets for the site of interest
#Grab a list of all Ameriflux sites, containing site ID and site description
sites_web <- jsonlite::fromJSON("http://ameriflux-data.lbl.gov/AmeriFlux/SiteSearch.svc/SiteList/AmeriFlux")
#Grab only NEON sites
sitesNeon <- sites_web[grep(pattern = paste0("NEON.*",site), x = sites_web$SITE_NAME),] #For all NEON sites
siteNeon <- sites_web[grep(pattern = paste0("NEON.*",site), x = sites_web$SITE_NAME),]
metaSite <- lapply(siteNeon$SITE_ID, function(x) {
pathSite <- paste0("http://ameriflux-data.lbl.gov/BADM/Anc/SiteInfo/",x)
tmp <- fromJSON(pathSite)
return(tmp)
}) Use Ameriflux site IDs to name metadata lists
#use NEON ID as list name
names(metaSite) <- site
#Use Ameriflux site ID as list name
#names(metaSite) <- sitesNeon$SITE_ID Check if dateBgn is defined, if not make it the initial operations date “IOCR” of the site
if(!exists("dateBgn") || is.na(dateBgn) || is.null(dateBgn)){
dateBgn <- as.Date(metaSite[[site]]$values$GRP_FLUX_MEASUREMENTS[[1]]$FLUX_MEASUREMENTS_DATE_START, "%Y%m%d")
} else {
dateBgn <- dateBgn
}#End of checks for missing dateBgn
#Check if dateEnd is defined, if not make it the system date
if(!exists("dateEnd") || is.na(dateEnd) || is.null(dateEnd)){
dateEnd <- as.Date(Sys.Date())
} else {
dateEnd <- dateEnd
}#End of checks for missing dateEndGrab the UTC time offset from the Ameriflux API
timeOfstUtc <- as.integer(metaSite[[site]]$values$GRP_UTC_OFFSET[[1]]$UTC_OFFSET)Create the date sequence
setDate <- seq(from = as.Date(dateBgn), to = as.Date(dateEnd), by = "month")Start processing the site time range specified, verify that the site and date range are specified as intended
msg <- paste0("Starting Ameriflux FP standard conversion processing workflow for ", site, " for ", dateBgn, " to ", dateEnd)
print(msg)## [1] "Starting Ameriflux FP standard conversion processing workflow for KONZ for 2020-03-01 to 2020-05-31"Create output directory by checking if the download directory exists and create it if not
if(dir.exists(DirDnld) == FALSE) dir.create(DirDnld, recursive = TRUE)
#Append the site to the base output directory
DirOut <- paste0(DirOutBase, "/", siteNeon$SITE_ID)
#Check if directory exists and create if not
if(!dir.exists(DirOut)) dir.create(DirOut, recursive = TRUE)Download and extract data
#Initialize data List
dataList <- list()
#Read data from the API
dataList <- lapply(setDate, function(x) {
date <- stringr::str_extract(x, pattern = paste0("[0-9]{4}", "-", "[0-9]{2}"))
tryCatch(neonUtilities::zipsByProduct(dpID = dpID, site = site, startdate = date, enddate = date, package = "basic", savepath = DirDnld, check.size = FALSE), error=function(e) NULL)
files <- list.files(paste0(DirDnld, "/filesToStack00200"))
utils::unzip(paste0(DirDnld, "/filesToStack00200/", files[grep(pattern = paste0(site,".*.", date, ".*.zip"), x = files)]), exdir = paste0(DirDnld, "/filesToStack00200"))
files <- list.files(paste0(DirDnld, "/filesToStack00200"))
R.utils::gunzip(paste0(DirDnld, "/filesToStack00200/", files[grep(pattern = paste0(site, ".*.", date, ".*.h5.gz"), x = files)]), remove = FALSE)
files <- list.files(paste0(DirDnld, "/filesToStack00200"))
dataIdx <- rhdf5::h5read(file = paste0(DirDnld, "/filesToStack00200/", max(files[grep(pattern = paste0(site,".*.", date,".*.h5$"), x = files)])), name = paste0(site, "/"))
if(!is.null(dataIdx)){
dataIdx$dp0p <- NULL
dataIdx$dp02 <- NULL
dataIdx$dp03 <- NULL
dataIdx$dp01$ucrt <- NULL
dataIdx$dp04$ucrt <- NULL
dataIdx$dp01$data <- lapply(dataIdx$dp01$data,FUN=function(var){
nameTmi <- names(var)
var <- var[grepl('_30m',nameTmi)]
return(var)})
dataIdx$dp01$qfqm <- lapply(dataIdx$dp01$qfqm,FUN=function(var){
nameTmi <- names(var)
var <- var[grepl('_30m',nameTmi)]
return(var)})
}
return(dataIdx)
})## Finding available files
##
|
| | 0%
|
|======================================================================| 100%
##
## Downloading files totaling approximately 83.333588 MB
## Downloading 1 files
##
|
| | 0%
|
|======================================================================| 100%
## 1 files successfully downloaded to C:\Users\rohan\AppData\Local\Temp\RtmpCELr4B/filesToStack00200
## Finding available files
##
|
| | 0%
|
|======================================================================| 100%
##
## Downloading files totaling approximately 68.719186 MB
## Downloading 1 files
##
|
| | 0%
|
|======================================================================| 100%
## 1 files successfully downloaded to C:\Users\rohan\AppData\Local\Temp\RtmpCELr4B/filesToStack00200
## Finding available files
##
|
| | 0%
|
|======================================================================| 100%
##
## Downloading files totaling approximately 69.960912 MB
## Downloading 1 files
##
|
| | 0%
|
|======================================================================| 100%
## 1 files successfully downloaded to C:\Users\rohan\AppData\Local\Temp\RtmpCELr4B/filesToStack00200Add names to list for year/month combinations
names(dataList) <- paste0(lubridate::year(setDate),sprintf("%02d",lubridate::month(setDate)))Remove NULL elements from list
dataList <- dataList[vapply(dataList, Negate(is.null), NA)]Determine tower horizontal & vertical indices
#Find the tower top level by looking at the vertical index of the turbulent CO2 concentration measurements
LvlTowr <- grep(pattern = "_30m", names(dataList[[1]]$dp01$data$co2Turb), value = TRUE)
LvlTowr <- gsub(x = LvlTowr, pattern = "_30m", replacement = "")
#get tower top level
LvlTop <- strsplit(LvlTowr,"")
LvlTop <- base::as.numeric(LvlTop[[1]][6])
#Ameriflux vertical levels based off of https://ameriflux.lbl.gov/data/aboutdata/data-variables/ section 3.3.1 "Indices must be in order, starting with the highest."
idxVerAmfx <- base::seq(from = 1, to = LvlTop, by = 1)
#get the sequence from top to first level
LvlMeas <- base::seq(from = LvlTop, to = 1, by = -1)
#Recreate NEON naming conventions
LvlMeas <- paste0("000_0",LvlMeas,"0",sep="")
#Give NEON naming conventions to Ameriflux vertical levels
names(idxVerAmfx) <- LvlMeas
#Ameriflux horizontal index
idxHorAmfx <- 1Subset to the Ameriflux variables to convert
dataListFlux <- lapply(names(dataList), function(x) {
data.frame(
"TIMESTAMP_START" = as.POSIXlt(dataList[[x]]$dp04$data$fluxCo2$turb$timeBgn, format="%Y-%m-%dT%H:%M:%OSZ", tz = "GMT"),
"TIMESTAMP_END" = as.POSIXlt(dataList[[x]]$dp04$data$fluxCo2$turb$timeEnd, format="%Y-%m-%dT%H:%M:%OSZ", tz = "GMT"),
# "TIMESTAMP_START" = strftime(as.POSIXlt(dataList[[x]][[idxSite]]$dp04$data$fluxCo2$turb$timeBgn, format="%Y-%m-%dT%H:%M:%OSZ"), format = "%Y%m%d%H%M"),
# "TIMESTAMP_END" = strftime(as.POSIXlt(dataList[[x]][[idxSite]]$dp04$data$fluxCo2$turb$timeEnd, format="%Y-%m-%dT%H:%M:%OSZ") + 60, format = "%Y%m%d%H%M"),
"FC"= dataList[[x]]$dp04$data$fluxCo2$turb$flux,
"SC"= dataList[[x]]$dp04$data$fluxCo2$stor$flux,
"NEE"= dataList[[x]]$dp04$data$fluxCo2$nsae$flux,
"LE" = dataList[[x]]$dp04$data$fluxH2o$turb$flux,
"SLE" = dataList[[x]]$dp04$data$fluxH2o$stor$flux,
"USTAR" = dataList[[x]]$dp04$data$fluxMome$turb$veloFric,
"H" = dataList[[x]]$dp04$data$fluxTemp$turb$flux,
"SH" = dataList[[x]]$dp04$data$fluxTemp$stor$flux,
"FETCH_90" = dataList[[x]]$dp04$data$foot$stat$distXaxs90,
"FETCH_MAX" = dataList[[x]]$dp04$data$foot$stat$distXaxsMax,
"V_SIGMA" = dataList[[x]]$dp04$data$foot$stat$veloYaxsHorSd,
#"W_SIGMA" = dataList[[x]]$dp04$data$foot$stat$veloZaxsHorSd,
"CO2_1_1_1" = dataList[[x]]$dp01$data$co2Turb[[paste0(LvlTowr,"_30m")]]$rtioMoleDryCo2$mean,
"H2O_1_1_1" = dataList[[x]]$dp01$data$h2oTurb[[paste0(LvlTowr,"_30m")]]$rtioMoleDryH2o$mean,
"qfFinlH2oTurbFrt00Samp" = dataList[[x]]$dp01$qfqm$h2oTurb[[paste0(LvlTowr,"_30m")]]$frt00Samp$qfFinl,
"qfH2O_1_1_1" = dataList[[x]]$dp01$qfqm$h2oTurb[[paste0(LvlTowr,"_30m")]]$rtioMoleDryH2o$qfFinl,
"qfCO2_1_1_1" = dataList[[x]]$dp01$qfqm$co2Turb[[paste0(LvlTowr,"_30m")]]$rtioMoleDryCo2$qfFinl,
"qfSC" = dataList[[x]]$dp04$qfqm$fluxCo2$stor$qfFinl,
"qfSLE" = dataList[[x]]$dp04$qfqm$fluxH2o$stor$qfFinl,
"qfSH" = dataList[[x]]$dp04$qfqm$fluxTemp$stor$qfFinl,
"qfT_SONIC" = dataList[[x]]$dp01$qfqm$soni[[paste0(LvlTowr,"_30m")]]$tempSoni$qfFinl,
"qfWS_1_1_1" = dataList[[x]]$dp01$qfqm$soni[[paste0(LvlTowr,"_30m")]]$veloXaxsYaxsErth$qfFinl,
rbind.data.frame(lapply(names(idxVerAmfx), function(y) {
tryCatch({rlog$debug(y)}, error=function(cond){print(y)})
rpt <- list()
rpt[[paste0("CO2_1_",idxVerAmfx[y],"_2")]] <- dataList[[x]]$dp01$data$co2Stor[[paste0(y,"_30m")]]$rtioMoleDryCo2$mean
rpt[[paste0("H2O_1_",idxVerAmfx[y],"_2")]] <- dataList[[x]]$dp01$data$h2oStor[[paste0(y,"_30m")]]$rtioMoleDryH2o$mean
rpt[[paste0("CO2_1_",idxVerAmfx[y],"_3")]] <- dataList[[x]]$dp01$data$isoCo2[[paste0(y,"_30m")]]$rtioMoleDryCo2$mean
rpt[[paste0("H2O_1_",idxVerAmfx[y],"_3")]] <- dataList[[x]]$dp01$data$isoCo2[[paste0(y,"_30m")]]$rtioMoleDryH2o$mean
rpt[[paste0("qfCO2_1_",idxVerAmfx[y],"_2")]] <- dataList[[x]]$dp01$qfqm$co2Stor[[paste0(LvlTowr,"_30m")]]$rtioMoleDryCo2$qfFinl
rpt[[paste0("qfH2O_1_",idxVerAmfx[y],"_2")]] <- dataList[[x]]$dp01$qfqm$h2oStor[[paste0(LvlTowr,"_30m")]]$rtioMoleDryH2o$qfFinl
rpt[[paste0("qfCO2_1_",idxVerAmfx[y],"_3")]] <- dataList[[x]]$dp01$qfqm$isoCo2[[paste0(LvlTowr,"_30m")]]$rtioMoleDryCo2$qfFinl
rpt[[paste0("qfH2O_1_",idxVerAmfx[y],"_3")]] <- dataList[[x]]$dp01$qfqm$isoH2o[[paste0(LvlTowr,"_30m")]]$rtioMoleDryH2o$qfFinl
rpt <- rbind.data.frame(rpt)
return(rpt)
}
)),
"WS_1_1_1" = dataList[[x]]$dp01$data$soni[[paste0(LvlTowr,"_30m")]]$veloXaxsYaxsErth$mean,
"WS_MAX_1_1_1" = dataList[[x]]$dp01$data$soni[[paste0(LvlTowr,"_30m")]]$veloXaxsYaxsErth$max,
"WD_1_1_1" = dataList[[x]]$dp01$data$soni[[paste0(LvlTowr,"_30m")]]$angZaxsErth$mean,
"T_SONIC" = dataList[[x]]$dp01$data$soni[[paste0(LvlTowr,"_30m")]]$tempSoni$mean,
"T_SONIC_SIGMA" = base::sqrt(dataList[[x]]$dp01$data$soni[[paste0(LvlTowr,"_30m")]]$tempSoni$mean)
, stringsAsFactors = FALSE)
})## [1] "000_040"
## [1] "000_030"
## [1] "000_020"
## [1] "000_010"## Warning in base::sqrt(dataList[[x]]$dp01$data$soni[[paste0(LvlTowr, "_30m")]]
## $tempSoni$mean): NaNs produced## [1] "000_040"
## [1] "000_030"
## [1] "000_020"
## [1] "000_010"## Warning in base::sqrt(dataList[[x]]$dp01$data$soni[[paste0(LvlTowr, "_30m")]]
## $tempSoni$mean): NaNs produced## [1] "000_040"
## [1] "000_030"
## [1] "000_020"
## [1] "000_010" names(dataListFlux) <- names(dataList)Combine the monthly data into a single dataframe, remove lists and clean memory
dataDfFlux <- do.call(rbind.data.frame,dataListFlux)
rm(list=c("dataListFlux","dataList"))
gc()## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 1494506 79.9 3173284 169.5 3173284 169.5
## Vcells 16220600 123.8 72272750 551.4 75217183 573.9Regularize timeseries to 30 minutes in case timestamps are missing from NEON files due to processing errors
timeRglr <- eddy4R.base::def.rglr(timeMeas = as.POSIXlt(dataDfFlux$TIMESTAMP_START), dataMeas = dataDfFlux, BgnRglr = as.POSIXlt(dataDfFlux$TIMESTAMP_START[1]), EndRglr = as.POSIXlt(dataDfFlux$TIMESTAMP_END[length(dataDfFlux$TIMESTAMP_END)]), TzRglr = "UTC", FreqRglr = 1/(60*30))
#Reassign data to data.frame
dataDfFlux <- timeRglr$dataRglr
#Format timestamps
dataDfFlux$TIMESTAMP_START <- strftime(timeRglr$timeRglr + lubridate::hours(timeOfstUtc), format = "%Y%m%d%H%M")
dataDfFlux$TIMESTAMP_END <- strftime(timeRglr$timeRglr + lubridate::hours(timeOfstUtc) + lubridate::minutes(30), format = "%Y%m%d%H%M")Define validation times, and remove this data from the dataset. At NEON sites, validations with a series of gasses of known concentration are run every 23.5 hours. These values are used to correct for measurment drift and are run every 23.5 hours to achive daily resolution while also spreading the impact of lost measurements throughout the day.
#Remove co2Turb and h2oTurb data based off of qfFlow (qfFinl frt00)
dataDfFlux$FC[(which(dataDfFlux$qfCO2_1_1_1 == 1))] <- NaN
dataDfFlux$LE[(which(dataDfFlux$qfH2O_1_1_1 == 1))] <- NaN
dataDfFlux$USTAR[(which(dataDfFlux$qfWS_1_1_1 == 1))] <- NaN
dataDfFlux$H[(which(dataDfFlux$qfT_SONIC_1_1_1 == 1))] <- NaN
dataDfFlux$SC[(which(dataDfFlux$qfSC == 1))] <- NaN
dataDfFlux$SLE[(which(dataDfFlux$qfSLE == 1))] <- NaN
dataDfFlux$SH[(which(dataDfFlux$qfSH == 1))] <- NaN
dataDfFlux$T_SONIC[(which(dataDfFlux$qfT_SONIC_1_1_1 == 1))] <- NaN
dataDfFlux$T_SONIC_SIGMA[(which(dataDfFlux$qfT_SONIC_1_1_1 == 1))] <- NaN
dataDfFlux$WS_1_1_1[(which(dataDfFlux$qfWS_1_1_1 == 1))] <- NaN
dataDfFlux$WS_MAX_1_1_1[(which(dataDfFlux$qfWS_1_1_1 == 1))] <- NaN
dataDfFlux$WD_1_1_1[(which(dataDfFlux$qfWS_1_1_1 == 1))] <- NaN
dataDfFlux$H2O_1_1_1[(which(dataDfFlux$qfH2O_1_1_1 == 1))] <- NaN
dataDfFlux$CO2_1_1_1[(which(dataDfFlux$qfCO2_1_1_1 == 1))] <- NaN
lapply(idxVerAmfx, function(x){
#x <- 1
dataDfFlux[[paste0("H2O_1_",x,"_2")]][(which(dataDfFlux[[paste0("qfH2O_1_",x,"_2")]] == 1))] <<- NaN
dataDfFlux[[paste0("H2O_1_",x,"_3")]][(which(dataDfFlux[[paste0("qfH2O_1_",x,"_3")]] == 1))] <<- NaN
dataDfFlux[[paste0("CO2_1_",x,"_2")]][(which(dataDfFlux[[paste0("qfCO2_1_",x,"_2")]] == 1))] <<- NaN
dataDfFlux[[paste0("CO2_1_",x,"_3")]][(which(dataDfFlux[[paste0("qfCO2_1_",x,"_3")]] == 1))] <<- NaN
})## $`000_040`
## [1] NaN
##
## $`000_030`
## [1] NaN
##
## $`000_020`
## [1] NaN
##
## $`000_010`
## [1] NaNRemove quality flagging variables from output
setIdxQf <- grep("qf", names(dataDfFlux))
dataDfFlux[,setIdxQf] <- NULLSet range thresholds
#assign list
Rng <- list()
Rng$Min <- data.frame(
"FC" = -100, #[umol m-2 s-1]
"SC" = -100, #[umol m-2 s-1]
"NEE" = -100, #[umol m-2 s-1]
"LE" = -500, #[W m-2]
"H" = -500, #[W m-2]
"USTAR" = 0, #[m s-1]
"CO2" = 200, #[umol mol-1]
"H2O" = 0, #[mmol mol-1]
"WS_1_1_1" = 0, #[m s-1]
"WS_MAX_1_1_1" = 0, #[m s-1]
"WD_1_1_1" = -0.1, #[deg]
"T_SONIC" = -55.0 #[C]
)Set Max thresholds
Rng$Max <- data.frame(
"FC" = 100, #[umol m-2 s-1]
"SC" = 100, #[umol m-2 s-1]
"NEE" = 100, #[umol m-2 s-1]
"LE" = 1000, #[W m-2]
"H" = 1000, #[W m-2]
"USTAR" = 5, #[m s-1]
"CO2" = 800, #[umol mol-1]
"H2O" = 100, #[mmol mol-1]
"WS_1_1_1" = 50, #[m s-1]
"WS_MAX_1_1_1" = 50, #[m s-1]
"WD_1_1_1" = 360, #[deg]
"T_SONIC" = 45.0 #[C]
)Grab all CO2/H2O columns to apply same thresholds, replace missing values with -9999
nameCO2 <- grep("CO2",names(dataDfFlux),value = TRUE)
nameH2O <- grep("H2O",names(dataDfFlux),value = TRUE)
#Apply the CO2/H2O threshold to all variables in HOR_VER_REP
Rng$Min[nameCO2] <- Rng$Min$CO2
Rng$Min[nameH2O] <- Rng$Min$H2O
Rng$Max[nameCO2] <- Rng$Max$CO2
Rng$Max[nameH2O] <- Rng$Max$H2O
#Apply the range test to the output, and replace values with NaN
lapply(names(dataDfFlux), function(x) {
dataDfFlux[which(dataDfFlux[,x]<Rng$Min[[x]] | dataDfFlux[,x]>Rng$Max[[x]]),x] <<- NaN})## [[1]]
## [1] NaN
##
## [[2]]
## [1] NaN
##
## [[3]]
## [1] NaN
##
## [[4]]
## [1] NaN
##
## [[5]]
## [1] NaN
##
## [[6]]
## [1] NaN
##
## [[7]]
## [1] NaN
##
## [[8]]
## [1] NaN
##
## [[9]]
## [1] NaN
##
## [[10]]
## [1] NaN
##
## [[11]]
## [1] NaN
##
## [[12]]
## [1] NaN
##
## [[13]]
## [1] NaN
##
## [[14]]
## [1] NaN
##
## [[15]]
## [1] NaN
##
## [[16]]
## [1] NaN
##
## [[17]]
## [1] NaN
##
## [[18]]
## [1] NaN
##
## [[19]]
## [1] NaN
##
## [[20]]
## [1] NaN
##
## [[21]]
## [1] NaN
##
## [[22]]
## [1] NaN
##
## [[23]]
## [1] NaN
##
## [[24]]
## [1] NaN
##
## [[25]]
## [1] NaN
##
## [[26]]
## [1] NaN
##
## [[27]]
## [1] NaN
##
## [[28]]
## [1] NaN
##
## [[29]]
## [1] NaN
##
## [[30]]
## [1] NaN
##
## [[31]]
## [1] NaN
##
## [[32]]
## [1] NaN
##
## [[33]]
## [1] NaN
##
## [[34]]
## [1] NaN
##
## [[35]]
## [1] NaN
##
## [[36]]
## [1] NaN # Delete any NEE that have either FC or SC removed
dataDfFlux[is.na(dataDfFlux$FC) | is.na(dataDfFlux$SC),"NEE"] <- NaN
#Change NA to -9999
dataDfFlux[is.na(dataDfFlux)] <- -9999Write output data to csv
#Create output filename based off of Ameriflux file naming convention
nameFileOut <- base::paste0(DirOut,"/",siteNeon$SITE_ID,'_HH_',dataDfFlux$TIMESTAMP_START[1],'_',utils::tail(dataDfFlux$TIMESTAMP_END,n=1),'_flux.csv')
#Write output to .csv
write.csv(x = dataDfFlux, file = nameFileOut, row.names = FALSE)Clean up environment
rm(list="dataDfFlux")
gc()## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 1496962 80.0 3173284 169.5 3173284 169.5
## Vcells 16294743 124.4 57818200 441.2 75217183 573.9## [1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE5.7 Flux Measurements and Interoperability Exercise
5.7.1 NEON Flux Coding Lab
- NEON data are submitted to AmeriFlux quarterly after one year of non-quality flagged or otherwise missing data are available. Use the workflow above to extend the data coverage of an already submitted NEON site by downloading existing data from the AmeriFlux website and recently published HDF5 files from the NEON data portal. Process the NEON data such that it is in AmeriFlux format and plot the entire timerseries.
Hint: NEON sites start with US-x
- Using metScanR package, find co-located NEON and AmeriFlux sites. Download data for an overlapping time period, and compare FC and H values by making a scatter plot and seeing how far off the data are from a 1:1 line.